Team:NC School of Sci Math/Lab Notebook
From 2013hs.igem.org
Home Team Project Details Lab Notebook Results Human Impact Biosafety Acknowledgments Official Team Profile
Planning and Development
01/11/2013
Brainstorming for project ideas- LDL-cholesterol biosensor
- Ocean salinity regulator (especially for coral reefs)
- Water contaminant multibiosensor
More research should be done
- Find related projects that have been completed
- Obstacles?
- BioBricks?
- Presentations in 3-4 weeks
01/18/2013
Discussions of each project- LDL-cholesterol biosensor
- Could be used as a blood test
- Blood sugar monitor-like device
- Hard to measure any color change in blood
- Ocean salinity regulator (especially for coral reefs)
- Need a lot of bacteria
- Environmental concerns
- Bacteria can do well in coral reefs
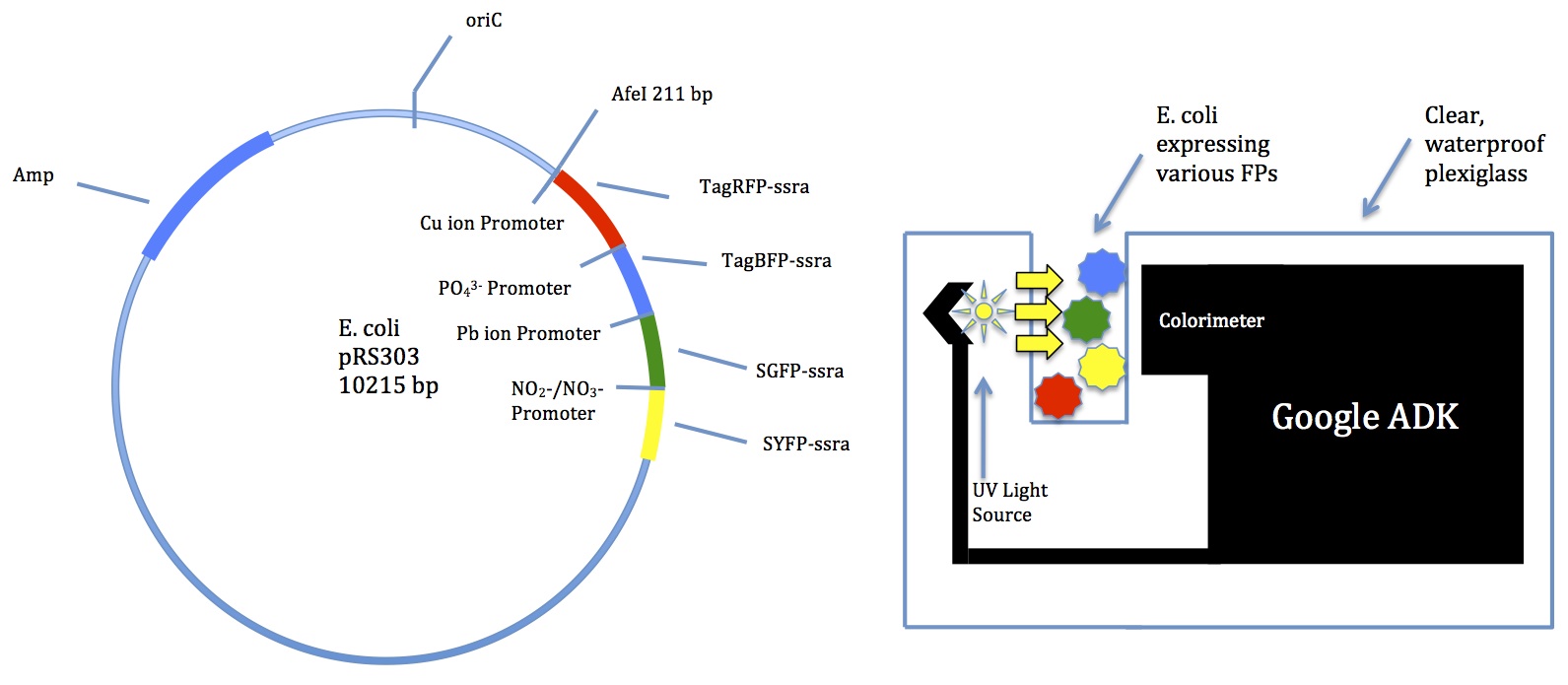
- Water contaminant multibiosensor
- Several Biobrick promoters for common contaminants
- Usage in sewer tanks
- Many different colors of fluorescent proteins
- Could incorporate newer technology for better color-sensing
- Will look up data on promoter/FP expression
02/01/2013
Presentations
- All ideas sound promising
- Meet next week to decide which idea to pursue
02/08/2013
Decision on project- LDL-cholesterol idea won't work
- Costly and hard to detect FPs in blood
- Multibiosensor is appealing
- Many applications
- Expandable with many different FPs
- Could bring in new technology
- Ocean salinity regulator is difficult because of the ocean
- Environmental factors and unknown variables
Multibiosensor chosen
02/15/2013
A few members met with Mr. Jon Davis to discuss potential technology
- Suggested a Google device called ADK
- Comes with a colorimeter
- Can be programmed for many functions
- School has a few we could try out
02/22/2013
Goals for the immediate future
- Find Biobricks
- Set up schedule for lab work
- Further research into possible uses
- Preliminary lab experiments
Project Proposal
03/01/2013
Discussion of upcoming project proposal:
- Introduction, Solution Statement, and Methods
- Need to find out some background information
- Should make use of Tinkercell modeling
03/04/2013
Assignments of roles for the upcoming project proposal:
- Madeline will investigate background information
- Danny will work on Tinkercell models
- Jack will write the solution statement and methods sections
- Synthesize our writing on a Google Doc
- Try to have individual parts done in about 2 weeks
03/18/2013
The following models have been created
- We should use these in the proposal, but continue to work on the Tinkercell model
03/25/2013
Everyone began editing the proposal on a Google Doc today
- Want to be finished by March 29th
- Tinkercells will probably not be ready in time
- Having a lot of trouble getting fluorescent protein levels to zero when they should be
- Danny will talk to Morgan to see if he can help
- Edit Edit Edit!
03/27/2013
- Proposal is coming along well
- Need to finish the methods, but otherwise complete
- Take pictures of the ADK and lab work
03/29/2013
- Proposal is done!
- Will submit soon
04/06/2013
Time to focus on creating the plasmid
- Finished Tinkercell model will help a bunch
- Do we want to think about ordering de novo from a gene synthesis company?
- Computer modeling is the best approach for now
04/18/2013
- Danny has a working Tinkercell model for the copper promoter!
- We should be able to figure out the others soon
- The lead detector may be especially difficult because the ion binds to a promoter to create a transcription factor
Colony PCR:
Screening of the two different biobrick clonings done by colony-PCR.
Protocol:
A reaction mix was prepared containing 0.2 µl of each screening primer used for the screening, 9.6 µl of water and 10 µl of 2x PCR mastermix (fermentas). Finally, a colony was picked from a plate using a sterile pipette tip and was dipped into the PCR mix a few times. 12 clones were screened for each different cloning setup as follows:
precB_LacZ: primers VF2: and precB Reverse: were used. Only in case precB was successfully cloned into the backbone containing the reporter gene, we would get a PCR product.
precC_LacZ: primers VF2 and VR (standard sequencing primers) were used. We compared the product size of the different clones to the product size from the original vectors from the registry (only containing precA or LacZ). In case the cloning was successful, we should see a 200 bp shift in product size, which can be detected by gel electrophoresis.
The PCR program was done as follows:
94°C/3min
 "
"