Team:NC School of Sci Math/Project
From 2013hs.igem.org
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | <html><head> | ||
| + | <style type="text/css"> | ||
| + | #bodyContent h2, h2, #maincontent h2 | ||
| + | { margin-top: 0.5em; | ||
| + | margin-bottom: 0.65em; | ||
| + | font-size: 110%; | ||
| + | padding-bottom: 4px; } | ||
| + | </style> | ||
| + | </head></html> | ||
{{NCSSM_iGEM_13| | {{NCSSM_iGEM_13| | ||
Content= | Content= | ||
| Line 4: | Line 13: | ||
==Project Proposal== | ==Project Proposal== | ||
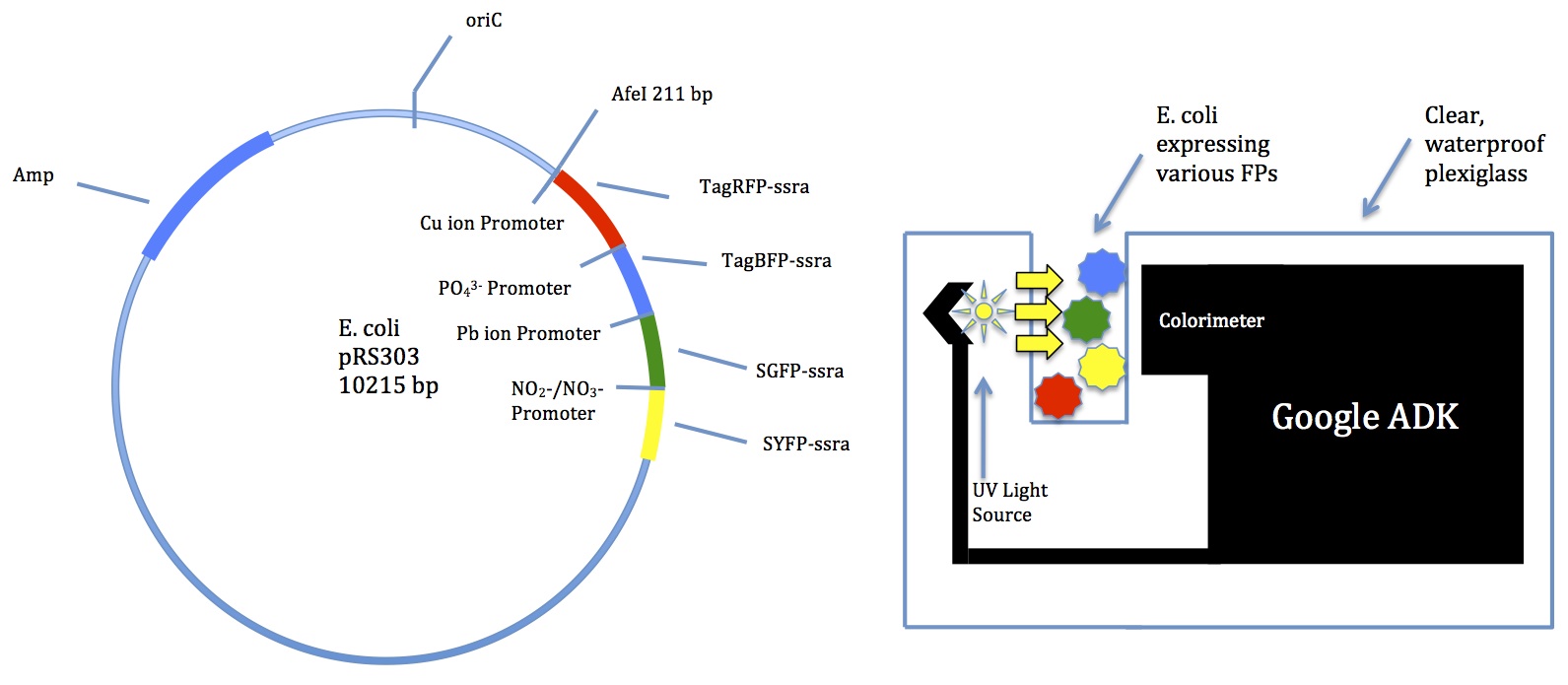
| - | Our multi-input logic gate will produce a colorimetric output for nitrates, phosphates, and toxic chemicals such as | + | Our multi-input logic gate will produce a colorimetric output for nitrates, phosphates, and toxic chemicals such as lead and copper, which are all detrimental to the function of septic tanks. The diagram below shows the genome of our E. Coli plasmid that will produce a unique colorimetric output based on the presence of the targeted metals in the local environment. |
| - | Figure 1 | + | As shown in Figure 1 on the left, prs303 E. coli plasmid after insertion of the genetic elements necessary for expression of fluorescent proteins based on presence of phosphates, nitrates, or reactive metal ions. Three of the four FPs in the plasmid are regulated by a different promoter that is activated in the presence of the targeted substance. Activation of the copper activated, phosphate activated and nitrate/nitrite activated promoters results in the translation of TagRFP, TagBFP2, and SYFP proteins, respectively. The fourth FP, Superfolder GFP, is regulated by a transcription factor that forms as the result of Lead ions binding to the Lead Binding Protein. The appropriate FP genes will be inserted at the AfeI restriction site, as shown. The Ampicillin Resistance Gene allows positive selection of plasmids that have been transformed as intended, and is inserted at the EcoRI site as shown. |
| - | Figure 2 | + | As shown in Figure 2 on the right, the Google ADK (Accessories Development Kit) can be used to sense these colorimetric outputs. The Google ADK can be modified to detect the colorimetric outputs from the E. coli plasmid. The device monitors color by detecting the amount and type of light absorbed across a known difference. A prototype of the device is shown in Figure 2. The chemicals present and their approximate concentrations based on absorbance of color are then sent through the Google ADK to the septic tank owner to notify them that an inspection of the tank is needed. If significant color change that could indicate increased levels of one or more of the harmful substances is detected, the modified Google ADK will notify the septic tank owner to recommend tank inspection. The Google ADK will serve as a detector of the colorimetric output and serves the main purpose of processing and communicating information to the tank owner. |
| + | [[File:Design.jpg]] | ||
| - | + | ==Methods== | |
| - | + | Initially, we started with the coding of the Google ADK. The goal was to program the ADK so that it outputs a text file containing the concentration of red, blue, and green light. After successful reprogramming, the ADK was tested with pGLO E. coli and different color lights to test the sensitivity of the ADK in detection of fluorescence protein and colored light. These tests proved to be successful when comparing the light intensity with and without the presence of pGLO E. coli. In addition, it was also noted that the color yellow increased the intensity of red and green light. With the ADK coded, we moved on to the design of the genetic network. | |
| + | The genetic network incorporates four biosensors created through existing bioparts in order to create one new biopart, which is the multibiosensor. Existing parts were selected from the Registry of Biological Parts to create each biosensor. The reporters were picked based on the intensity of light produced. The ssRA degradation tag was also used in each biosensor to decrease the time of the protein. | ||
| + | After creating each biosensor, restriction enzyme sites were attached to the ends of the biosensors so that they could be ligated together into the plasmid. NEB Cutter 2.0 was used to make sure the restriction enzymes used did not disrupt the coding sequence of the biosensor. If no overlap between possible restriction enzymes occurred, the restriction site sequence was attached to the ends the biosensors for future ligation. The location of the multibiosensor insert within the prs303 plasmid was confirmed as well. Next, ApE (A plasmid Editor) was used to construct the plasmid with the complete sequence. | ||
| + | |||
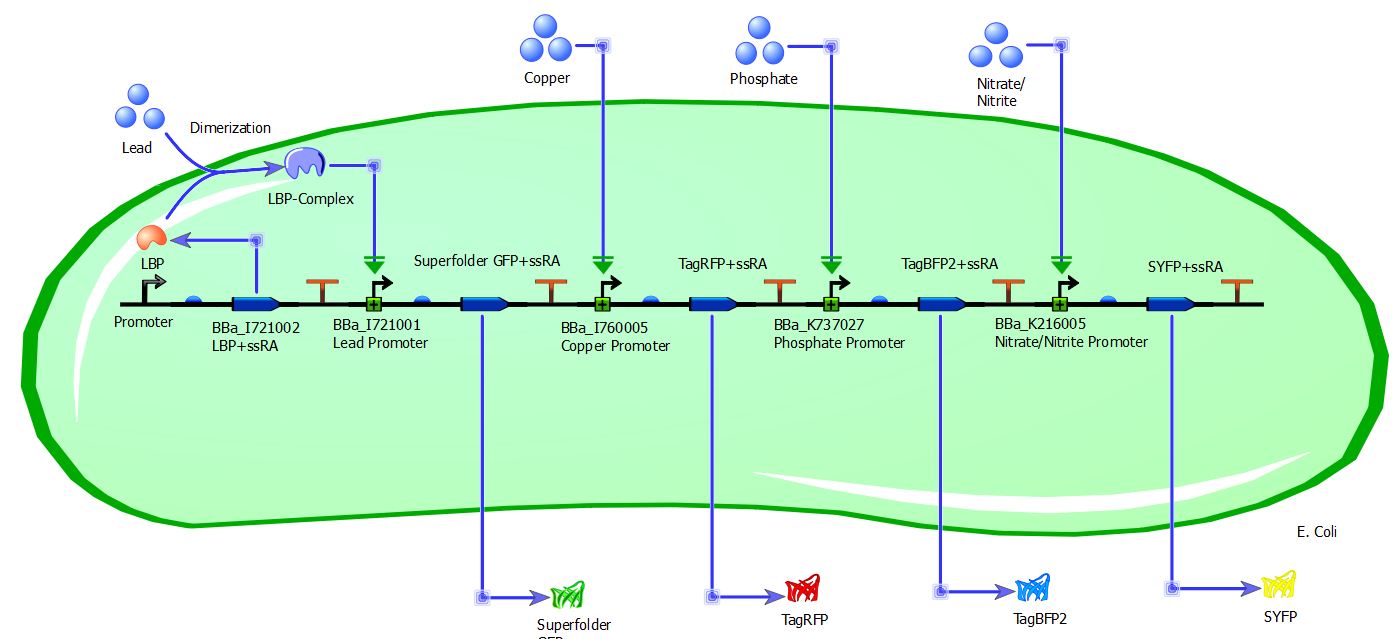
| + | Tinker Cell, a computer-aided design program for synthetic biology, was used to design the multibiosensor network. As shown by the model, the inducible promoter is followed by a ribosome binding site, coding sequence of the reporter with the ssRA degradation tag, and the transcriptional terminator. The activation of the inducible promoter by the ions and the production of the different fluorescent proteins are shown by the model as well. Lead provided an interesting case, in which the promoter could only be activated through the synthesis of the Lead Binding Protein Complex. This is also diagrammed in the model as well. With the genetic network, computational results were executed through the deterministic and stochastic simulation models. | ||
| + | |||
| + | [[File:model.jpg]] | ||
}} | }} | ||
Latest revision as of 03:55, 22 June 2013
Home Team Project Details Lab Notebook Results Human Impact Biosafety Acknowledgments Official Team Profile
Project Proposal
Our multi-input logic gate will produce a colorimetric output for nitrates, phosphates, and toxic chemicals such as lead and copper, which are all detrimental to the function of septic tanks. The diagram below shows the genome of our E. Coli plasmid that will produce a unique colorimetric output based on the presence of the targeted metals in the local environment.
As shown in Figure 1 on the left, prs303 E. coli plasmid after insertion of the genetic elements necessary for expression of fluorescent proteins based on presence of phosphates, nitrates, or reactive metal ions. Three of the four FPs in the plasmid are regulated by a different promoter that is activated in the presence of the targeted substance. Activation of the copper activated, phosphate activated and nitrate/nitrite activated promoters results in the translation of TagRFP, TagBFP2, and SYFP proteins, respectively. The fourth FP, Superfolder GFP, is regulated by a transcription factor that forms as the result of Lead ions binding to the Lead Binding Protein. The appropriate FP genes will be inserted at the AfeI restriction site, as shown. The Ampicillin Resistance Gene allows positive selection of plasmids that have been transformed as intended, and is inserted at the EcoRI site as shown.
As shown in Figure 2 on the right, the Google ADK (Accessories Development Kit) can be used to sense these colorimetric outputs. The Google ADK can be modified to detect the colorimetric outputs from the E. coli plasmid. The device monitors color by detecting the amount and type of light absorbed across a known difference. A prototype of the device is shown in Figure 2. The chemicals present and their approximate concentrations based on absorbance of color are then sent through the Google ADK to the septic tank owner to notify them that an inspection of the tank is needed. If significant color change that could indicate increased levels of one or more of the harmful substances is detected, the modified Google ADK will notify the septic tank owner to recommend tank inspection. The Google ADK will serve as a detector of the colorimetric output and serves the main purpose of processing and communicating information to the tank owner.
Methods
Initially, we started with the coding of the Google ADK. The goal was to program the ADK so that it outputs a text file containing the concentration of red, blue, and green light. After successful reprogramming, the ADK was tested with pGLO E. coli and different color lights to test the sensitivity of the ADK in detection of fluorescence protein and colored light. These tests proved to be successful when comparing the light intensity with and without the presence of pGLO E. coli. In addition, it was also noted that the color yellow increased the intensity of red and green light. With the ADK coded, we moved on to the design of the genetic network.
The genetic network incorporates four biosensors created through existing bioparts in order to create one new biopart, which is the multibiosensor. Existing parts were selected from the Registry of Biological Parts to create each biosensor. The reporters were picked based on the intensity of light produced. The ssRA degradation tag was also used in each biosensor to decrease the time of the protein.
After creating each biosensor, restriction enzyme sites were attached to the ends of the biosensors so that they could be ligated together into the plasmid. NEB Cutter 2.0 was used to make sure the restriction enzymes used did not disrupt the coding sequence of the biosensor. If no overlap between possible restriction enzymes occurred, the restriction site sequence was attached to the ends the biosensors for future ligation. The location of the multibiosensor insert within the prs303 plasmid was confirmed as well. Next, ApE (A plasmid Editor) was used to construct the plasmid with the complete sequence.
Tinker Cell, a computer-aided design program for synthetic biology, was used to design the multibiosensor network. As shown by the model, the inducible promoter is followed by a ribosome binding site, coding sequence of the reporter with the ssRA degradation tag, and the transcriptional terminator. The activation of the inducible promoter by the ions and the production of the different fluorescent proteins are shown by the model as well. Lead provided an interesting case, in which the promoter could only be activated through the synthesis of the Lead Binding Protein Complex. This is also diagrammed in the model as well. With the genetic network, computational results were executed through the deterministic and stochastic simulation models.
 "
"