Team:Beijing HDFLS High/Project
From 2013hs.igem.org
(→SOS RESPONSE) |
(→DNA Sequencing and Submitting Part) |
||
| (110 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<div id="header">{{:Team:Beijing_HDFLS_High/header}}</div> | <div id="header">{{:Team:Beijing_HDFLS_High/header}}</div> | ||
| - | == | + | == TITLE: The UV-responsive Wintergreen Odor Generator== |
| - | The | + | == ABSTRACT == |
| - | + | The UV-responsive wintergreen odor generator catalyzes the conversion of the precursor salicylic acid to the wintergreen odor methyl salicylate under extensive UV irradiation. The biosynthetic device is composed of two transcriptional devices: Rec A (SOS) promoter (BBa_J22106) and a wintergreen odor enzyme generator (BBa_J45119). The odor generator takes as input extensive UV irradiation and produce as output an enzyme that catalyzes production of an odor from a chemical precursor. | |
| - | + | [[Image:Generator.JPG|center|800px]] | |
| - | + | ===INTRODUCTION=== | |
| - | + | We designed a biological system using ''E.coli'' to detect extensive UV irradiation. If the toolkit is under extensive UV irradiation, it will produce a wintergreen odor.Then, This special odor will warn people of the UV irradiation. In order to increase the attraction of our product, we make ''E.coli'' culture medium with the different color using red, yellow and blue pigments.<br><br> | |
| - | We | + | [[Image:Outlook.jpg||Center||400px|]]<Br> |
| - | In | + | <br> |
| + | <br> | ||
| + | Ultraviolet (UV) light is part of the electromagnetic spectrum having waves with frequencies higher than those that humans can identify as the color violet. These frequencies are invisible to humans, but visible to a number of insects and birds.<br> | ||
| + | |||
| + | UVC | ||
| + | The shortest one of UV rays ,usually do not penetrate the Earth’s ozone layer. | ||
| + | <br> | ||
| + | UVB | ||
| + | Longer waves that reach skin’s surface – can cause surface tanning burning ,and signs of aging. | ||
| + | <br> | ||
| + | UVA | ||
| + | Even longer waves that can penetrate deep into the skin’s surface ,releasing free radicals and causing DNA changes that can result in skin cancers.<br> | ||
| - | + | [[File:uv.jpg||center||800px|]] | |
| - | + | <br>Recently, because the ozone layer has been destroying by some artificial chemical substances like CFC (Freon),the increase of UV irradiation appears on the earth. | |
| - | + | [[Image:Beijing_HDFLS_High_ESA.jpg||center|||800px|]] | |
| - | + | <br> | |
| - | + | Extensive UV irradiation is harmful to human being, such as | |
| + | * 1) Weakening of the immune system | ||
| + | * 2) Increased risk of skin cancer, cataracts | ||
| + | * 3) Causing heavy damage to genes | ||
| - | == | + | == PARTS== |
| - | [[Image: | + | [[Image:1.jpg|center|700px]] |
| - | + | <br> | |
| + | Extensive UV irradiation impings on heavily DNA damage and results in a single-strand DNA occuring, which activates RecA protein.Then,RecA protein leads to the expression of about 15 different repair proteins to rescue DNA damage.RecA protein is the central regulator in SOS repair system.Rec A (SOS) Promoter is available in 2004 registry part plate. | ||
| + | [[Image:3.jpg|center|700px]] | ||
| + | BBa_J45119 takes as input a transcriptional signal (PoPS) and produce as output the BMST1 enzyme that catalyzes production of methyl salicylate from salicylic acid. Methyl salicylate has a wintergreen smell. Wintergreen odor enzyme (BMST1) generator is availabe in 2005 registry part plate. | ||
| + | [[Image:4.jpg|center|700px]] | ||
| - | == | + | == RESULTS == |
| - | === | + | === Growing Part A and Part B === |
| - | + | After Growing Part A After Growing Part B | |
| - | [[Image: | + | [[Image:浩泽菌落1.jpg|left|350px]] |
| - | + | [[Image:浩泽菌落2.jpg|right|350px]] | |
| + | <br> | ||
| + | <br> | ||
| + | <br><br> | ||
| + | <br> | ||
| + | <br><br> | ||
| + | <br> | ||
| + | <br><br> | ||
| + | <br> | ||
| + | <br><br> | ||
| + | <br> | ||
| + | <br><br><br> | ||
| + | <br> | ||
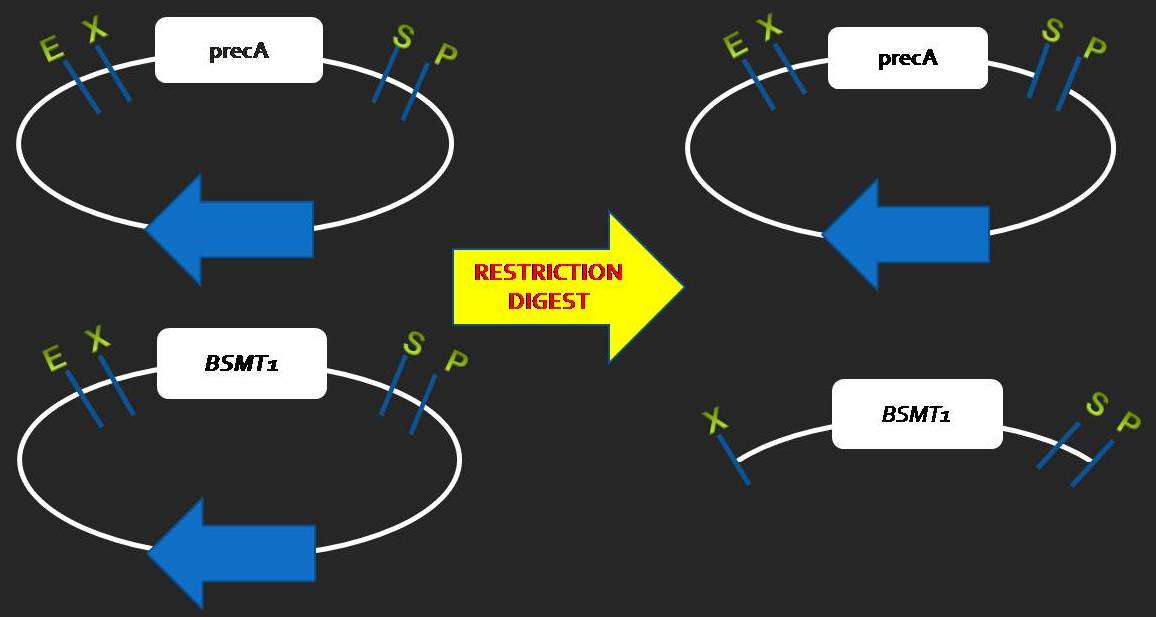
| - | == | + | === Resriction digest of Part A and Part B === |
| - | === STEP1: | + | [[Image:Digestion.jpg|center|500px]] |
| + | |||
| + | |||
| + | [[Image:PARTAandB.JPG|center|500px]] | ||
| + | |||
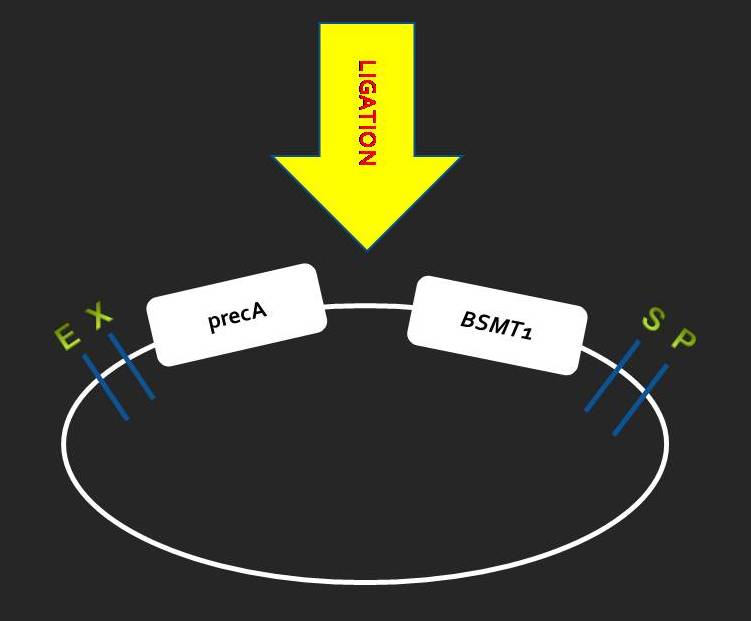
| + | === Growing After Ligation === | ||
| + | [[Image:ligation.jpg|center|310px]] | ||
| + | <br> | ||
| + | |||
| + | [[Image:lala.jpg|center|310px]] | ||
| + | |||
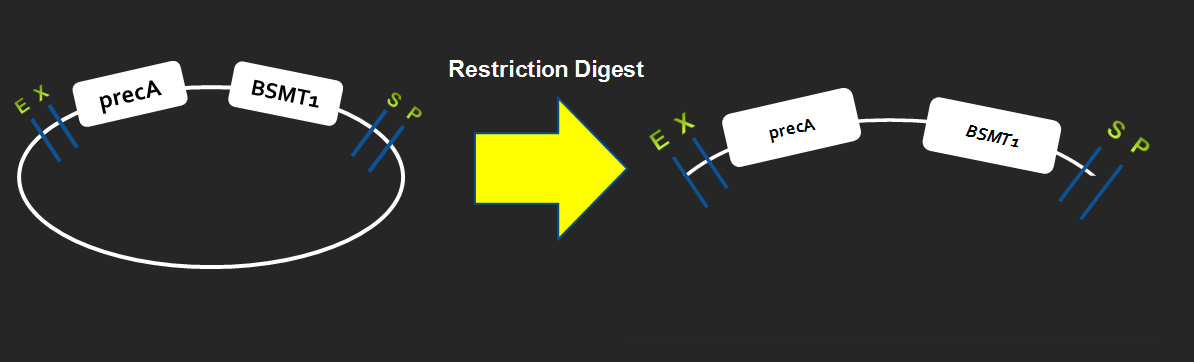
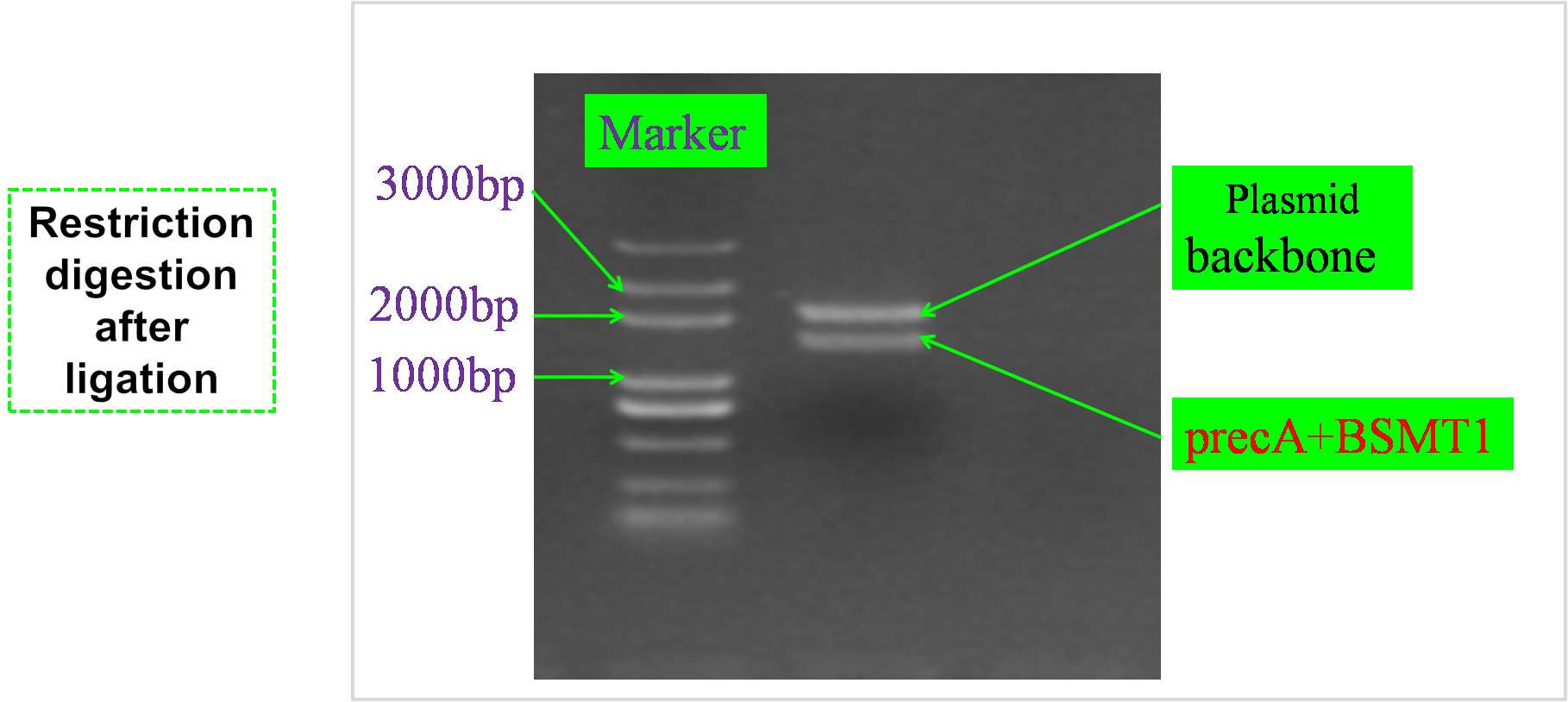
| + | === Restriction digest after ligation === | ||
| + | [[Image:AB.png|center|800px]] | ||
| + | |||
| + | |||
| + | [[Image:PARTAB.JPG|center|500px]] | ||
| + | |||
| + | === DNA Sequencing and Submitting Part=== | ||
| + | http://parts.igem.org/Part:BBa_K994000 | ||
| + | [[Image:Bases.jpg|center|800px]] | ||
| + | |||
| + | == STEP== | ||
| + | === STEP1: TRANSFORMATION AND GROWING === | ||
(Make sure we have enough copies of BBa_J22106 and BBa_J45119.) | (Make sure we have enough copies of BBa_J22106 and BBa_J45119.) | ||
| - | + | [[Image:Growing.jpg|center|600px]] | |
| - | + | === STEP2: Plamid MINIPREP === | |
| - | + | [[Image:PLASMID.jpg|center|600px]] | |
| - | + | ||
| - | + | === STEP3:RESTRICTION DIGEST === | |
| - | + | [[Image:Qiyue.JPG|center|600px]] | |
| - | + | <br> | |
| - | + | ||
| - | + | [[Image:20130518 102210.jpg|center|600px]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | === STEP2:MINIPREP === | + | |
| - | === STEP3:RESTRICTION DIGEST === | + | |
=== STEP4:LIGATION === | === STEP4:LIGATION === | ||
| + | [[Image:Menxuyang.jpg|center|600px]] | ||
=== STEP5:TRANSFORMATION === | === STEP5:TRANSFORMATION === | ||
| - | === | + | |
| - | === | + | === STEP6:MINIPREP === |
| + | === STEP7:RESTRICTION DIGEST === | ||
| + | === STEP8:DNA SEQUENCING=== | ||
| + | === STEP9:UV INDUCTION=== | ||
| + | === STEP10:DESGIN AND COMPLETE === | ||
| + | [[Image:Progect3.jpg|center|600px]] | ||
Latest revision as of 03:42, 22 June 2013
Contents |
TITLE: The UV-responsive Wintergreen Odor Generator
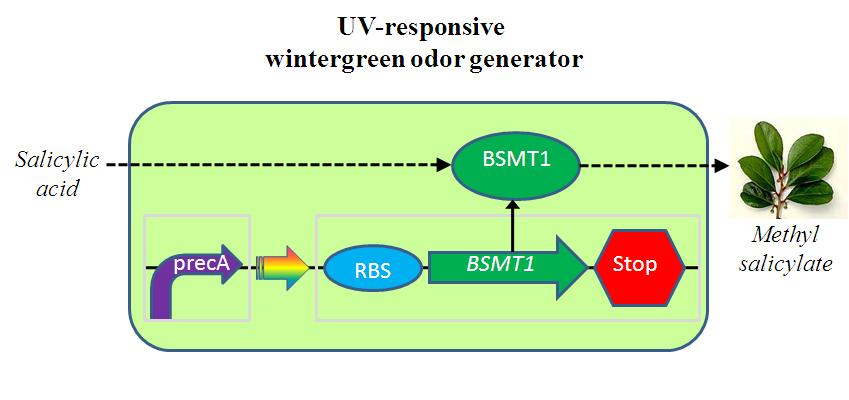
ABSTRACT
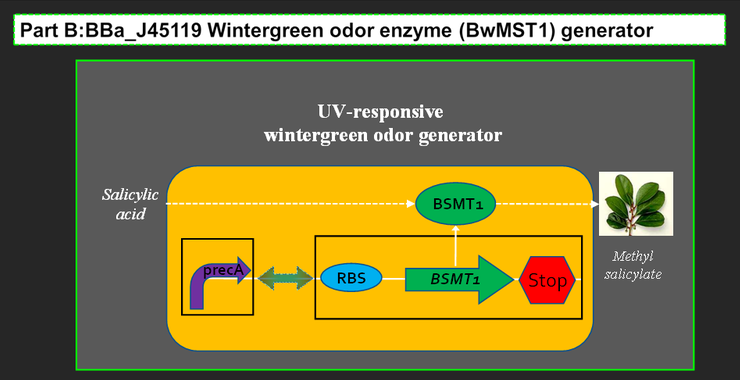
The UV-responsive wintergreen odor generator catalyzes the conversion of the precursor salicylic acid to the wintergreen odor methyl salicylate under extensive UV irradiation. The biosynthetic device is composed of two transcriptional devices: Rec A (SOS) promoter (BBa_J22106) and a wintergreen odor enzyme generator (BBa_J45119). The odor generator takes as input extensive UV irradiation and produce as output an enzyme that catalyzes production of an odor from a chemical precursor.
INTRODUCTION
We designed a biological system using E.coli to detect extensive UV irradiation. If the toolkit is under extensive UV irradiation, it will produce a wintergreen odor.Then, This special odor will warn people of the UV irradiation. In order to increase the attraction of our product, we make E.coli culture medium with the different color using red, yellow and blue pigments.

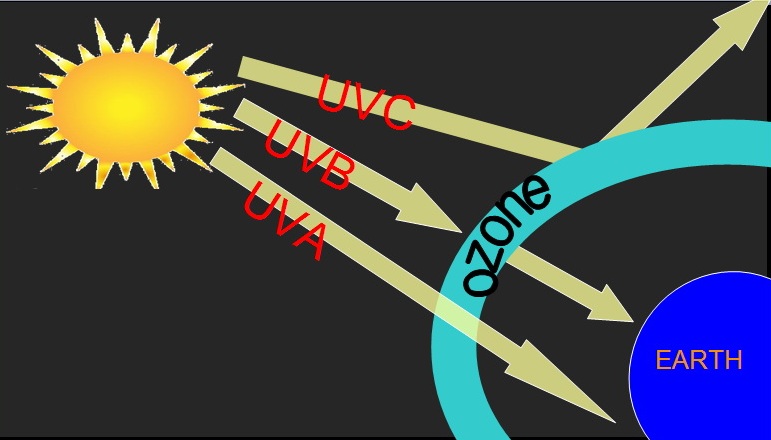
Ultraviolet (UV) light is part of the electromagnetic spectrum having waves with frequencies higher than those that humans can identify as the color violet. These frequencies are invisible to humans, but visible to a number of insects and birds.
UVC
The shortest one of UV rays ,usually do not penetrate the Earth’s ozone layer.
UVB
Longer waves that reach skin’s surface – can cause surface tanning burning ,and signs of aging.
UVA
Even longer waves that can penetrate deep into the skin’s surface ,releasing free radicals and causing DNA changes that can result in skin cancers.
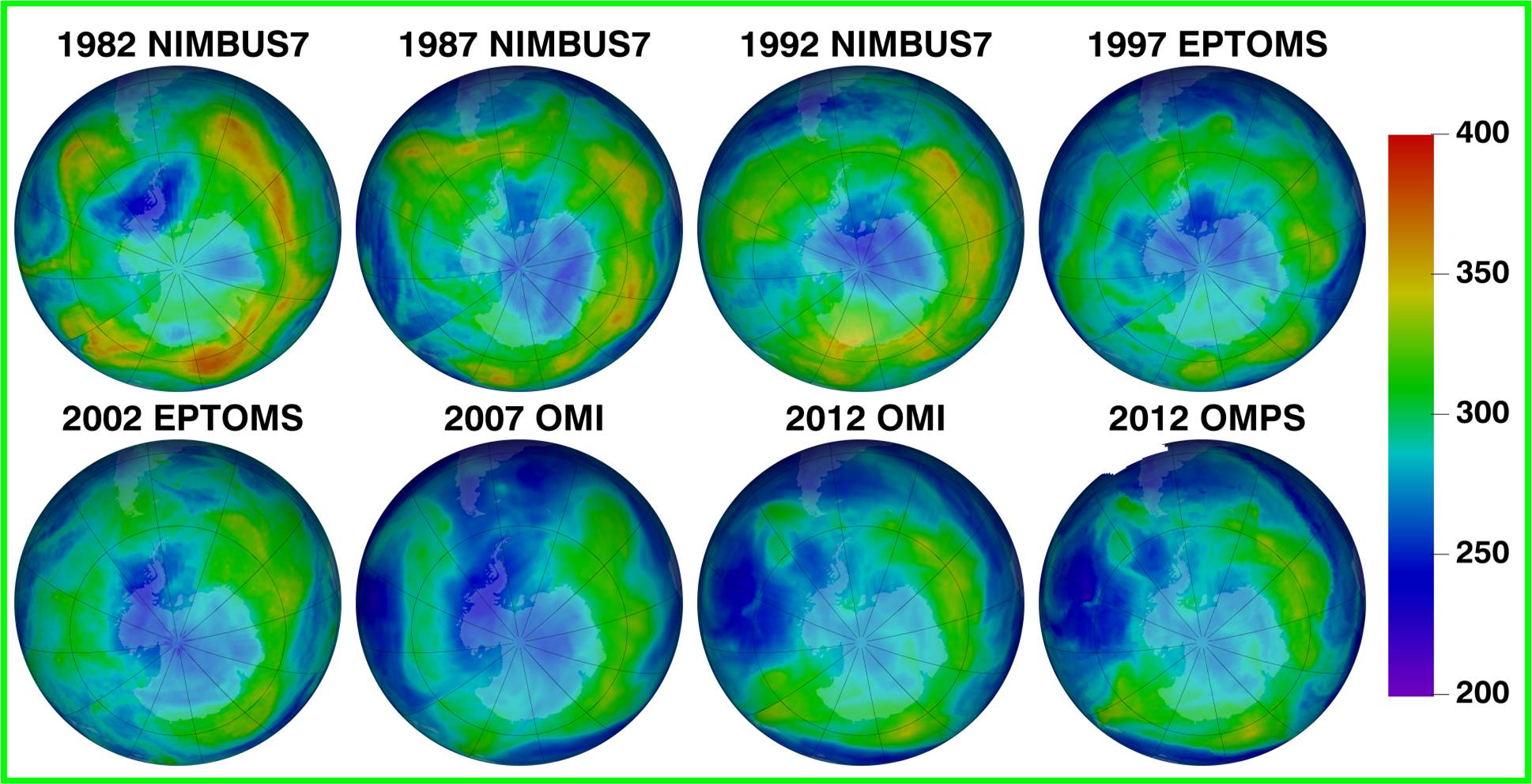
Recently, because the ozone layer has been destroying by some artificial chemical substances like CFC (Freon),the increase of UV irradiation appears on the earth.
Extensive UV irradiation is harmful to human being, such as
- 1) Weakening of the immune system
- 2) Increased risk of skin cancer, cataracts
- 3) Causing heavy damage to genes
PARTS
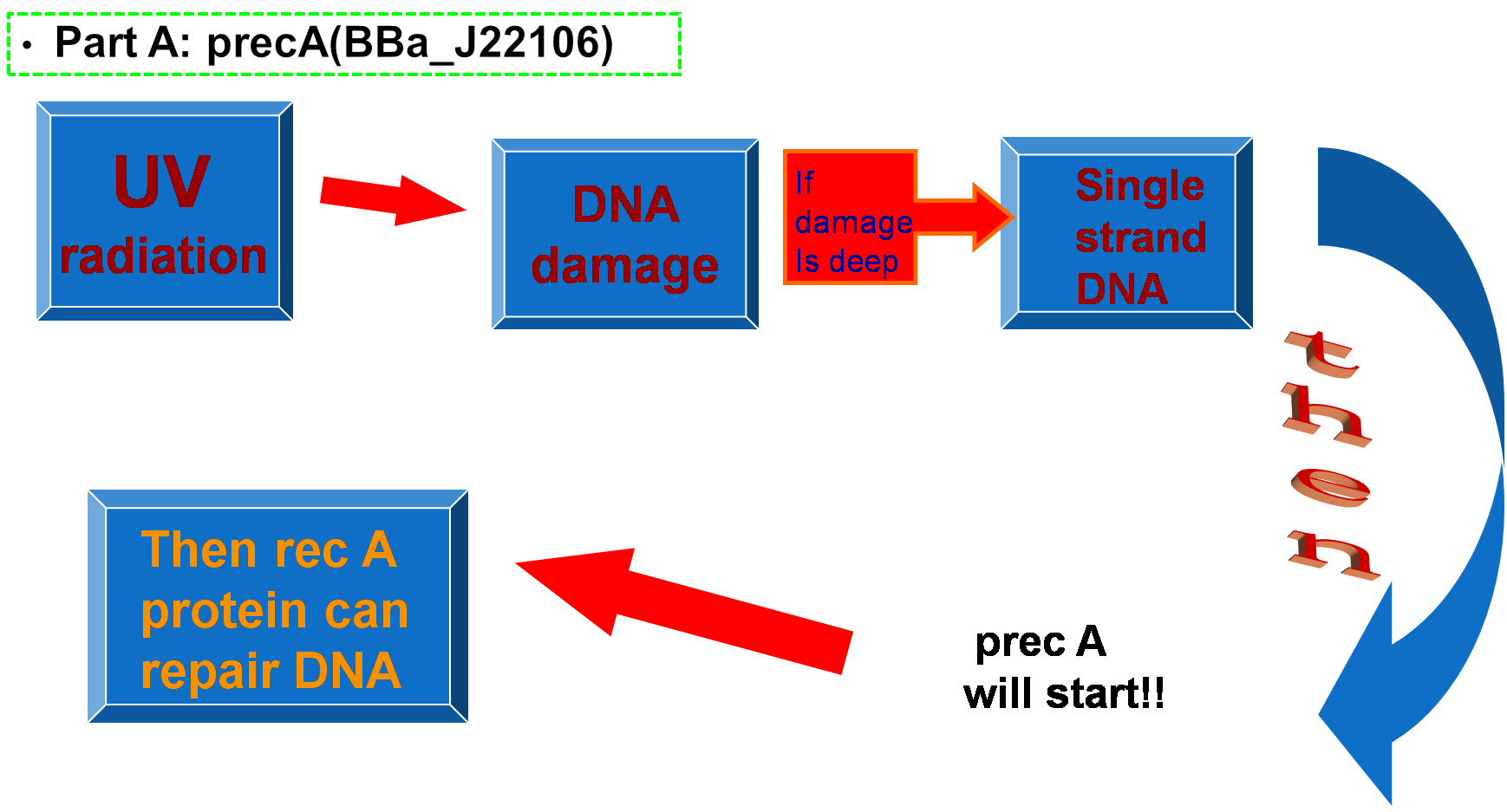
Extensive UV irradiation impings on heavily DNA damage and results in a single-strand DNA occuring, which activates RecA protein.Then,RecA protein leads to the expression of about 15 different repair proteins to rescue DNA damage.RecA protein is the central regulator in SOS repair system.Rec A (SOS) Promoter is available in 2004 registry part plate.
BBa_J45119 takes as input a transcriptional signal (PoPS) and produce as output the BMST1 enzyme that catalyzes production of methyl salicylate from salicylic acid. Methyl salicylate has a wintergreen smell. Wintergreen odor enzyme (BMST1) generator is availabe in 2005 registry part plate.
RESULTS
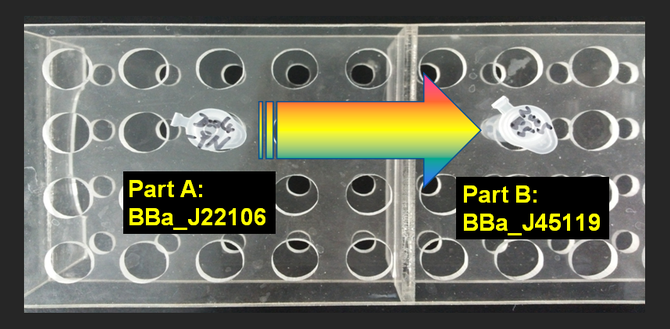
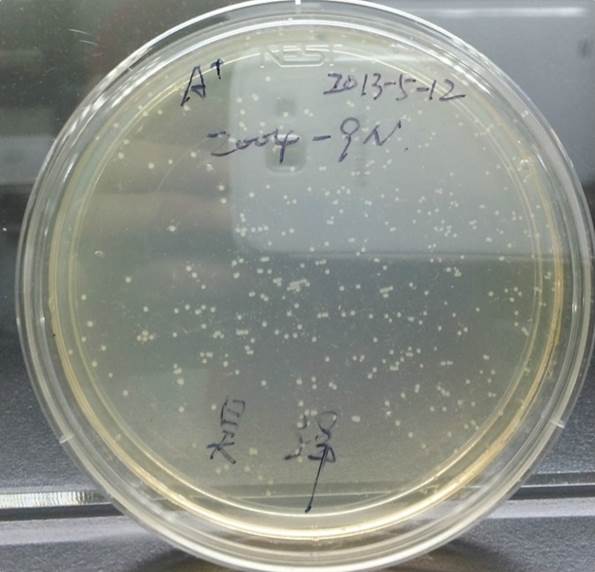
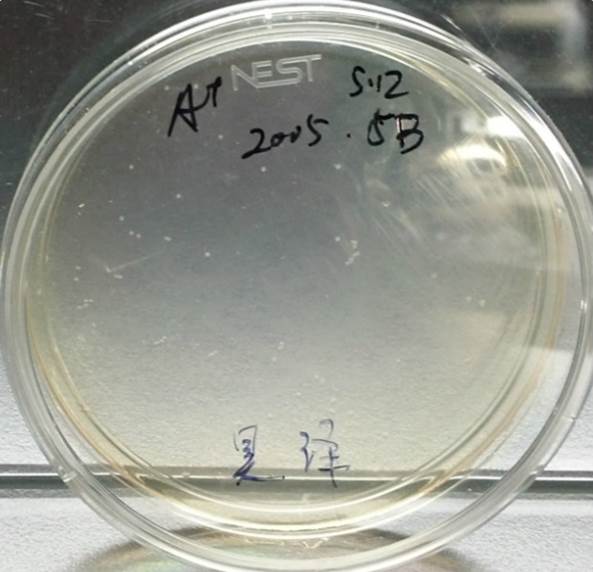
Growing Part A and Part B
After Growing Part A After Growing Part B
Resriction digest of Part A and Part B
Growing After Ligation
Restriction digest after ligation
DNA Sequencing and Submitting Part
http://parts.igem.org/Part:BBa_K994000
STEP
STEP1: TRANSFORMATION AND GROWING
(Make sure we have enough copies of BBa_J22106 and BBa_J45119.)
STEP2: Plamid MINIPREP
STEP3:RESTRICTION DIGEST
 "
"