Team:Beijing HDFLS High/Project
From 2013hs.igem.org
(→Enzyme digestion after ligation) |
|||
| Line 11: | Line 11: | ||
BBa_J45119 takes as input a transcriptional signal (PoPS) and produce as output the BMST1 enzyme that catalyzes production of methyl salicylate from salicylic acid. Methyl salicylate has a wintergreen smell. Wintergreen odor enzyme (BMST1) generator is availabe in 2005 registry part plate. | BBa_J45119 takes as input a transcriptional signal (PoPS) and produce as output the BMST1 enzyme that catalyzes production of methyl salicylate from salicylic acid. Methyl salicylate has a wintergreen smell. Wintergreen odor enzyme (BMST1) generator is availabe in 2005 registry part plate. | ||
[[Image:PARTS.JPG|center|600px]] | [[Image:PARTS.JPG|center|600px]] | ||
| - | |||
== RESULTS == | == RESULTS == | ||
=== Enzyme digestion of Part A and Part B === | === Enzyme digestion of Part A and Part B === | ||
| Line 17: | Line 16: | ||
=== Enzyme digestion after ligation === | === Enzyme digestion after ligation === | ||
[[Image:PARTAB.JPG|center|500px]] | [[Image:PARTAB.JPG|center|500px]] | ||
| - | + | == EXPERIMENTAL THEORY == | |
| - | == | + | to connect them together to completely the whole system. |
| - | + | == STEP== | |
| - | + | === STEP1: transformation and growing === | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
| - | === STEP1: | + | |
(Make sure we have enough copies of BBa_J22106 and BBa_J45119.) | (Make sure we have enough copies of BBa_J22106 and BBa_J45119.) | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== STEP2:MINIPREP === | === STEP2:MINIPREP === | ||
=== STEP3:RESTRICTION DIGEST === | === STEP3:RESTRICTION DIGEST === | ||
=== STEP4:LIGATION === | === STEP4:LIGATION === | ||
=== STEP5:TRANSFORMATION === | === STEP5:TRANSFORMATION === | ||
| - | === | + | === STEP6:MINIPREP === |
| - | === | + | === STEP7:RESTRICTION DIGEST === |
| + | === STEP8:DNA SEQUENCING=== | ||
| + | === STEP9:UV INDUCTION=== | ||
| + | === STEP10:DESGIN AND COMPLETE === | ||
Revision as of 15:09, 5 June 2013
Contents |
ABSTRACT
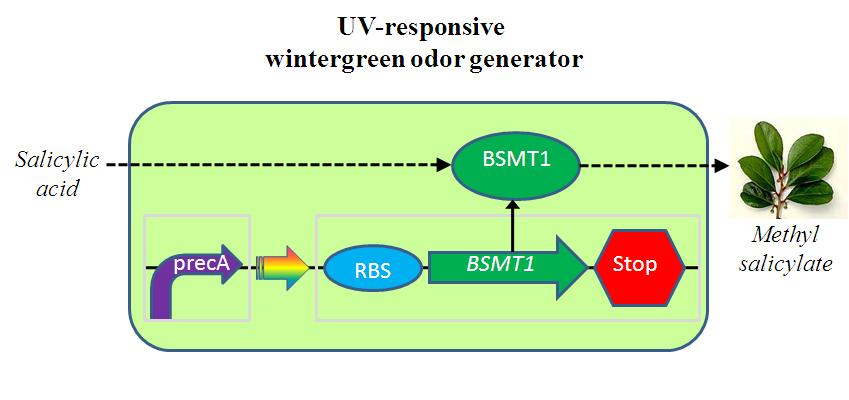
The UV-responsive wintergreen odor generator catalyzes the conversion of the precursor salicylic acid to the wintergreen odor methyl salicylate under extensive UV irradiation. The biosynthetic device is composed of two transcriptional devices: Rec A (SOS) promoter (BBa_J22106) and a wintergreen odor enzyme generator (BBa_J45119). The odor generator takes as input extensive UV irradiation and produce as output an enzyme that catalyzes production of an odor from a chemical precursor.
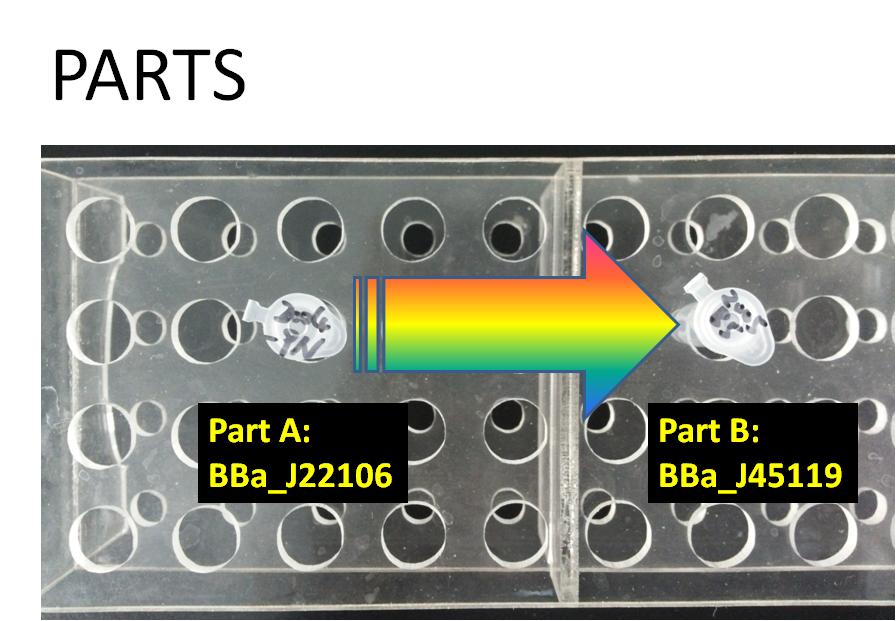
PARTS
Part A:BBa_J22106 rec A (SOS) Promoter
Extensive UV irradiation impings on heavily DNA damage, resulting in a single-strand DNA occuring, which activates RecA protein.Then,RecA protein leads to the expression of about 20 different repair proteins to contract DNA damamage.RecA protein is the central regulator in SOS repair system.Rec A (SOS) Promoter is available in 2004 registry part plate.
Part B:BBa_J45119 Wintergreen odor enzyme (BMST1) generator
BBa_J45119 takes as input a transcriptional signal (PoPS) and produce as output the BMST1 enzyme that catalyzes production of methyl salicylate from salicylic acid. Methyl salicylate has a wintergreen smell. Wintergreen odor enzyme (BMST1) generator is availabe in 2005 registry part plate.
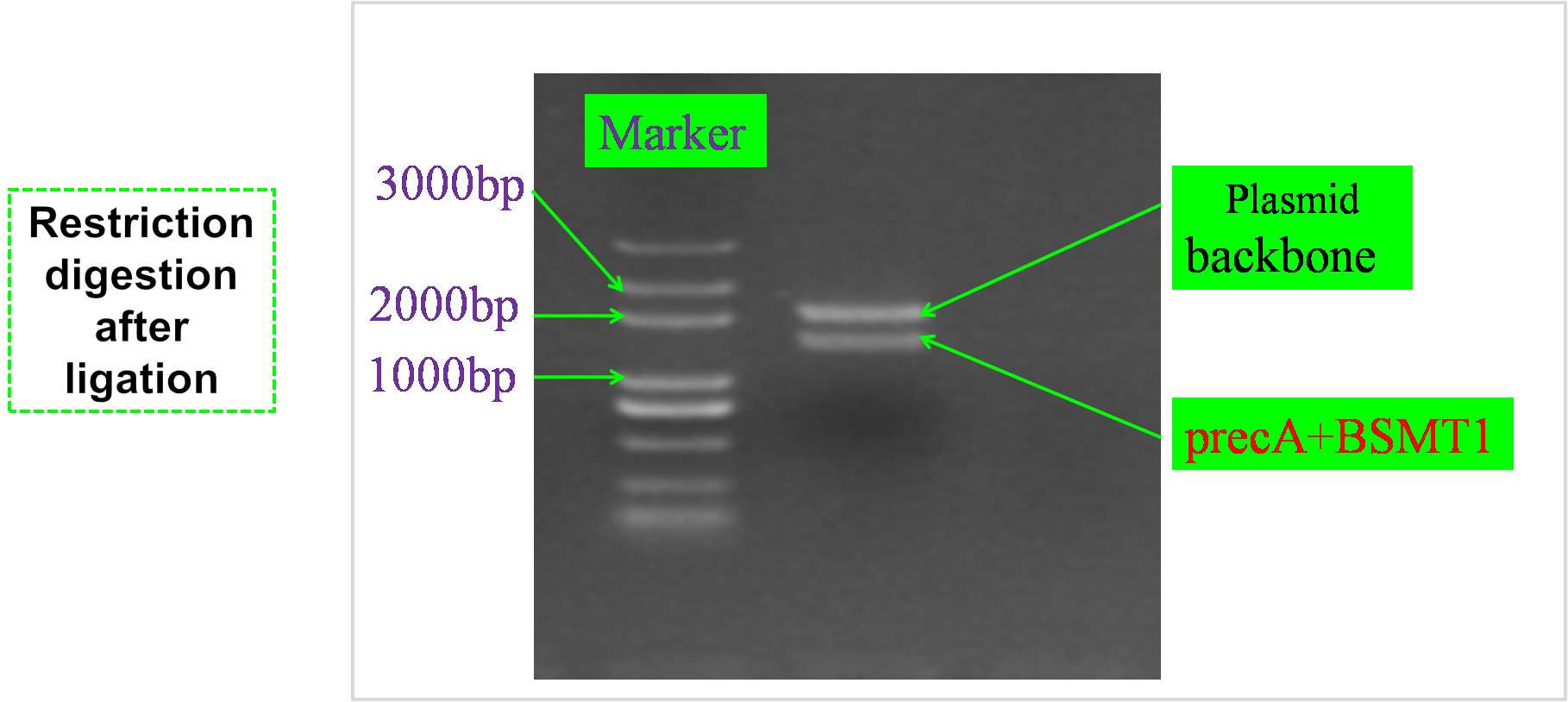
RESULTS
Enzyme digestion of Part A and Part B
Enzyme digestion after ligation
EXPERIMENTAL THEORY
to connect them together to completely the whole system.
STEP
STEP1: transformation and growing
(Make sure we have enough copies of BBa_J22106 and BBa_J45119.)
 "
"