Team:Beijing HDFLS High/Project
From 2013hs.igem.org
Contents |
ABSTRACT
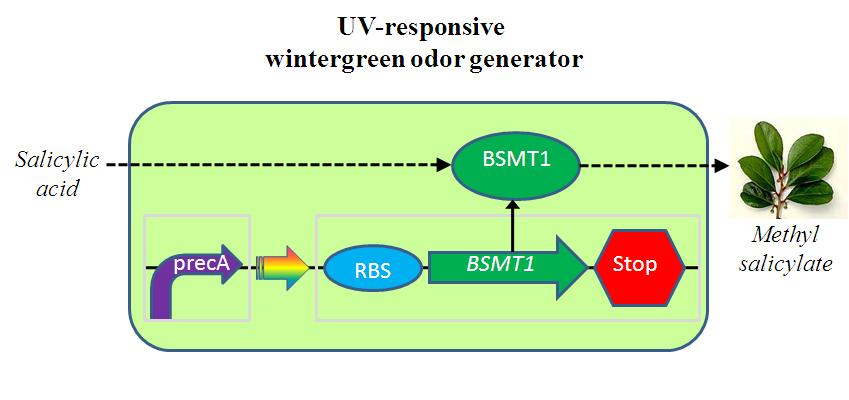
The UV-responsive wintergreen odor generator catalyzes the conversion of the precursor salicylic acid to the wintergreen odor methyl salicylate under extensive UV irradiation. The biosynthetic device is composed of two transcriptional devices: Rec A (SOS) promoter (BBa_J22106) and a wintergreen odor enzyme generator (BBa_J45119). The odor generator takes as input extensive UV irradiation and produce as output an enzyme that catalyzes production of an odor from a chemical precursor.
Parts
Parts
Promoter :RecA(BBa_J22106)
e chose to start with the promoter RecA , RecA is a recombinase enzyme, i.e. a DNA strand exchange and recombination protein with protease and nuclease activity. The RecA protein binds strongly and in long clusters to ssDNA to form a nucleoprotein filament. The protein has more than one DNA binding site, and thus can hold a single strand and double strand together. This feature makes it possible to catalyze a DNA synapsis reaction between a DNA double helix and a complementary region of single stranded DNA. And ReaA is the standard gene for the SOS-response therefore it is well characterized. Other scientist have been successful when using recA for gene regulation.
SOS RESPONSE
The SOS response was discovered and named by Miroslav Radman in 1975.The SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis are induced. The SOS system is the repair system of the cell to counteract extensive DNA damages e.g. caused by UV radiation or ionizing radiation. If UV radiation impinges on the bacterial DNA it may cause cross-linking of adjacent cytosine and thymine bases resulting in pyrimidine dimers – a process called direct DNA damage. Heavily damaged DNA is no longer able to replicate and leads to interruption of the cell cycle. The SOS response allows bacterial cells to repair damaged DNA and continue the cell cycle. In normal cells, the SOS system is turned off and it is only activated in the case of DNA damage. The central regulator of the SOS response is RecA. The RecA protein is activated by single-stranded DNA occurring during heavy DNA damage. Activated RecA leads to the cleavage of the LexA-repressor protein that normally blocks the RNA-polymerase for transcription of repair proteins. The inactivated LexA can no longer bind to the DNA strain which allows for the production of a cascade of about 20 different repair proteins, amongst others SulA, UvrB and DinF. Another action of the active LexA repressor protein is regulation of its own transcription as well as transcription of RecA . When DNA repair is done and the activity of precA has decreased the system returns to its steady state.
REPORTER : "Wintergreen odor enzyme (BMST1) generator" (BBa_J45119)
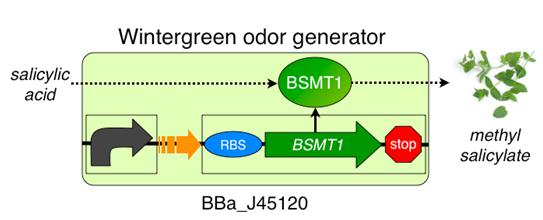
The wintergreen odor generator (BBa_J45120) catalyzes the conversion of the precursor salicylic acid to the odor methyl salicylate that has a wintergreen smell. The biosynthetic device is composed of two transcriptional devices: a constitutive transcription source (BBa_R0040) and an odor enzyme generator (BBa_J45119). Odor enzyme generators take as input a transcriptional signal and produce as output an enzyme that catalyzes production of an odor from a chemical precursor.
Method
Experimental theory
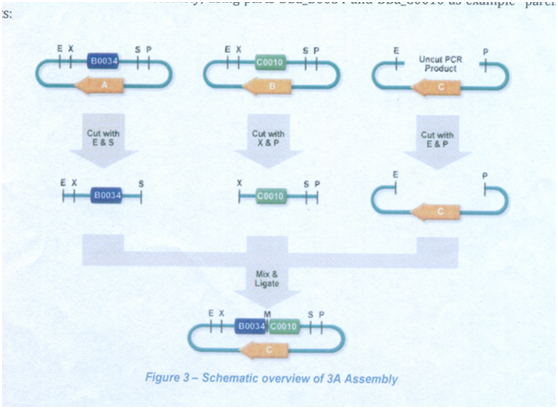
“3A(antibiotic)” Assembly is one way of assembling two strandard biological parts together. “3A(antibiotic)” Assembly uses the restriction sites in the prefix and suffix of the two parts to assemble them into the new composite part. The composite part will have the same prefix and suffix as the ‘parent’ parts.
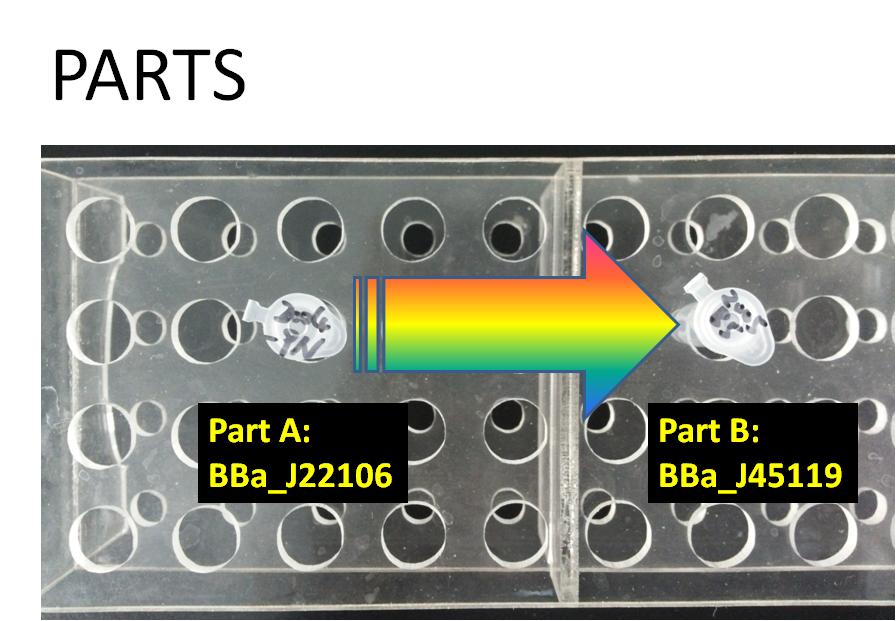
This time choose RecA as promoter and wintergreen odor biosynthetic system as reporter. The uesful part of RecA are cut out by enzymese ECORI and PSTI ,and we also need ECORI and PSTI to cut the useful part of wintergreen odor biosynthetic system ,and then we need to connect them together to completely the whole system.
Step
STEP1: GROWING
(Make sure we have enough copies of BBa_J22106 and BBa_J45119.)
- PART1:
- Clean the lab bench
- Label the agar plates with names of partA and parB and partC (partA:J22106 partB:J45119 partC: BBa_J45199banana)
- Use an inoculating loop to transfer some cells from the PartA agar stab to the appropriate agar plate .Dip an inoculating loop into the stab at the same location, and streak the bacteria onto the agar plate in a zig-zag pattern
- Repeat step 3 for partB and partC
- Place the agar plates into the incubator with the agar facing down. Incubate the agar plates at 37 degrees for 14-16hours. Alternately, incubate at room temperature for 24-30 hours.
- PART2
- Clean the lab bench
- Remove the agar plates from fridge
- Add 5ml of LB broth to each culture tube(with 14ml )
- Use an inoculating loop to pick a single colony from each agar plate and sent it into the culture tube.
- Incubate for 16 hours at 37 degrees in a shaker or rotator.
- Seal until use
 "
"