|
|
| (37 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| | + | <html><head> |
| | + | <style type="text/css"> |
| | + | |
| | + | h3 |
| | + | { font-color: #00008B; } |
| | + | |
| | + | #bodyContent h2, h2, #maincontent h2 |
| | + | { margin-top: 1em; |
| | + | margin-bottom: 0.95em; |
| | + | font-size: 110%; |
| | + | padding-bottom: 4px; } |
| | + | |
| | + | #maincontent ul, #maincontent li |
| | + | { list-style-type:square; |
| | + | list-style-image:url('https://2013hs.igem.org/wiki/skins/igem/bullet.gif'); } |
| | + | |
| | + | #maincontent ul |
| | + | { margin: 0px 22px; } |
| | + | |
| | + | </style> |
| | + | </head></html>{{NotebookUpper}} |
| | {{NCSSM_iGEM_13| | | {{NCSSM_iGEM_13| |
| | Content= | | Content= |
| | __NOTOC__ | | __NOTOC__ |
| - |
| |
| | <!-- | | <!-- |
| | | | |
| Line 15: |
Line 35: |
| | | | |
| | /--> | | /--> |
| - | {{NotebookUpper}}
| + | ===Lab Notebook=== |
| - | <html> | + | <html><h2><a name="Planning" style="color:#00008B;">Planning and Development</a></h2> |
| - | <style type="text/css">
| + | <h4><a style="color:#00008B; float:right;" name="01/11/2013">01/11/2013</a></h4></html> |
| - | h3 {font-color: #00008B;}
| + | |
| - | </style>
| + | |
| - | <div id="Scrollbereich">
| + | |
| - | <h2><a name="Planning" style="color:#00008B;">Planning and Development</a></h2> | + | |
| - | <h4><a style="color:#00008B;" name="01/11/2013">01/11/2013</a></h4> | + | |
| - | </html> | + | |
| | <b>Brainstorming for project ideas </b> | | <b>Brainstorming for project ideas </b> |
| | <br /> | | <br /> |
| Line 29: |
Line 44: |
| | *Ocean salinity regulator (especially for coral reefs) | | *Ocean salinity regulator (especially for coral reefs) |
| | *Water contaminant multibiosensor | | *Water contaminant multibiosensor |
| - | <br />
| + | |
| - | <b>More research should be done</b> | + | <b style="margin-top:2px;">More research</b> |
| | *Find related projects that have been completed | | *Find related projects that have been completed |
| | *Obstacles? | | *Obstacles? |
| Line 36: |
Line 51: |
| | *Presentations in 3-4 weeks | | *Presentations in 3-4 weeks |
| | | | |
| - | <html> | + | <html><h4><a style="color:#00008B; float:right;" name="01/18/2013">01/18/2013</a></h4></html> |
| - | <h4><a style="color:#00008B;" name="01/18/2013">01/18/2013</a></h4> | + | |
| - | </html> | + | |
| | <b>Discussions of each project</b> | | <b>Discussions of each project</b> |
| | <br /> | | <br /> |
| Line 56: |
Line 70: |
| | **Will look up data on promoter/FP expression | | **Will look up data on promoter/FP expression |
| | | | |
| - | <html> | + | <html><h4><a style="color:#00008B; float:right;" name="02/01/2013">02/01/2013</a></h4></html> |
| - | <h4><a style="color:#00008B;" name="02/01/2013">02/01/2013</a></h4> | + | |
| - | </html> | + | |
| | | | |
| | <b>Presentations</b> | | <b>Presentations</b> |
| Line 64: |
Line 76: |
| | *Meet next week to decide which idea to pursue | | *Meet next week to decide which idea to pursue |
| | | | |
| - | <html> | + | <html><h4><a style="color:#00008B; float:right;" name="02/08/2013">02/08/2013</a></h4></html> |
| - | <h4><a style="color:#00008B;" name="02/08/2013">02/08/2013</a></h4> | + | |
| - | </html> | + | |
| | <b>Decision on project</b> | | <b>Decision on project</b> |
| | *LDL-cholesterol idea won't work | | *LDL-cholesterol idea won't work |
| Line 74: |
Line 85: |
| | **Expandable with many different FPs | | **Expandable with many different FPs |
| | **Could bring in new technology | | **Could bring in new technology |
| - | *Ocean salinity regulator is difficult because of the ocean | + | *Ocean salinity regulator is difficult because of the ocean environment |
| | **Environmental factors and unknown variables | | **Environmental factors and unknown variables |
| | <b>Multibiosensor chosen</b> | | <b>Multibiosensor chosen</b> |
| | | | |
| - | <html> | + | <html><h4><a style="color:#00008B; float:right;" name="02/15/2013">02/15/2013</a></h4></html> |
| - | <h4><a style="color:#00008B;" name="02/15/2013">02/15/2013</a></h4> | + | |
| - | </html> | + | |
| | | | |
| - | <b> A few members met with Mr. Jon Davis to discuss potential technology </b> | + | <b>Meeting with Mr. Jon Davis to discuss potential technology</b> |
| | *Suggested a Google device called ADK | | *Suggested a Google device called ADK |
| | **Comes with a colorimeter | | **Comes with a colorimeter |
| Line 88: |
Line 97: |
| | **School has a few we could try out | | **School has a few we could try out |
| | | | |
| - | <html> | + | <html><h4><a style="color:#00008B; float:right;" name="02/22/2013">02/22/2013</a></h4></html> |
| - | <h4><a style="color:#00008B;" name="02/22/2013">02/22/2013</a></h4> | + | |
| - | </html> | + | |
| | | | |
| - | <b>Goals for the immediate future</b> | + | <b>Goals for immediate future</b> |
| | *Find Biobricks | | *Find Biobricks |
| | *Set up schedule for lab work | | *Set up schedule for lab work |
| Line 98: |
Line 105: |
| | *Preliminary lab experiments | | *Preliminary lab experiments |
| | | | |
| | + | <html><h2><a name="Proposal" style="color:#00008B;">Project Proposal</a></h2> |
| | + | <h4><a style="color:#00008B; float:right;" name="03/01/2013">03/01/2013</a></h4></html> |
| | | | |
| - | <html>
| + | <b>Discussion of upcoming project proposal</b>: |
| - | <h2><a name="Proposal" style="color:#00008B;">Project Proposal</a></h2>
| + | |
| - | <br />
| + | |
| - | <h4><a style="color:#00008B;" name="03/01/2013">03/01/2013</a></h4>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | <p><b> Discussion of upcoming project proposal</b>:</p>
| + | |
| | * Introduction, Solution Statement, and Methods | | * Introduction, Solution Statement, and Methods |
| | * Need to find out some background information | | * Need to find out some background information |
| | * Should make use of Tinkercell modeling | | * Should make use of Tinkercell modeling |
| | | | |
| - | <html> | + | <html><h4><a style="color:#00008B; float:right;" name="03/04/2013">03/04/2013</a></h4></html> |
| - | <h4><a style="color:#00008B;" name="03/04/2013">03/04/2013</a></h4> | + | |
| - | </html> | + | |
| | | | |
| - | <p><b> Assignments of roles for the upcoming project proposal</b>:</p>
| + | <b>Assignments of roles for the upcoming project proposal</b>: |
| | * Madeline will investigate background information | | * Madeline will investigate background information |
| | * Danny will work on Tinkercell models | | * Danny will work on Tinkercell models |
| Line 121: |
Line 122: |
| | * Try to have individual parts done in about 2 weeks | | * Try to have individual parts done in about 2 weeks |
| | | | |
| - | <html> | + | <html><h4><a style="color:#00008B; float:right;" name="03/18/2013">03/18/2013</a></h4></html> |
| - | <h4><a style="color:#00008B;" name="03/18/2013">03/18/2013</a></h4> | + | |
| - | </html> | + | |
| - | <p><b> The following models have been created</b>:</p>
| + | |
| - | [[File:Example.jpg]]
| + | |
| - | [[File:Example.jpg]]
| + | |
| - | | + | |
| - | <html>
| + | |
| - | <img src="https://static.igem.org/mediawiki/2012hs/3/3b/StdAss-edited.png" width=635>
| + | |
| - | </html><br/>
| + | |
| - | | + | |
| - | *<b>Gelelectrophoresis of the digested biobrick parts in order to extract the GFP and LacZ backbone as well as precA</b><br/>
| + | |
| - | <b>Protocol:</b>
| + | |
| - | In the meantime, the restriction digests were mixed with 6 µl of 6 x Loading Dye (fermentas). The whole restriction digest was loaded onto a 1 % agarose gel. Therefore, the agarose gel was prepared by adding 2 g of agarose to 200 ml of 1x TBE buffer and heated up in a microwave for 2 min/600 W. Thereby, the agarose was melted. Afterwards, the gel was stirred on a magnetic stirring device until the solution reached ~ 60°C and then the gel was poured. 10 µl of ethidium bromide were subsequently added by our supervisor Katharina Genreith (as EtBr is very toxic, we preferred our supervisor to do the handling of that when running a gel for the first time).
| + | |
| - | The samples were then loaded on the gel together with 1kb plus loading ladder (fermentas) and run for 1 h @ 100V. The expected bands can be seen on fig. 1.<br/>
| + | |
| - | <br/>
| + | |
| - | <html><img src="https://static.igem.org/mediawiki/2012hs/b/b7/Exp_gel.png" /><br/>
| + | |
| - | <b>Fig. 1: Schema of the gel we expect based on the length</b>(picture constructed using the WinSerial Cloner software)<br/>
| + | |
| - | <br/>
| + | |
| - | <img src="https://static.igem.org/mediawiki/2012hs/8/84/Gelelektrophorese.jpg" width ="500px" /><br/>
| + | |
| - | <b>Fig. 2: Picture taken of the gel.</b> All expected bands are visible<br/>
| + | |
| - | <br/>
| + | |
| - | </html>
| + | |
| - | *<b>Gel extraction of the indicated band (backbones for LacZ and GFP as well as precA insert) was done using a Qiagen gel extraction kit.</b><br/>
| + | |
| - | <b>Protocol:</b>
| + | |
| - | The bands were cut of the gel under UV light (caution: wear goggles in order to protect your eyes!) using a scalpel and the extracted band was put into a 2 ml eppi cup.<br/>
| + | |
| - | <br/><html>
| + | |
| - | <img src="https://static.igem.org/mediawiki/2012hs/a/aa/Gelelektrophoresenachher.jpg" width ="500px" /><br/>
| + | |
| - | <b>Fig. 3: Picture taken of the cut gel.</b> This picture was taken after the required bands were excised for gel extraction<br/>
| + | |
| - | <br/></html>
| + | |
| - | Afterwards, 600 µl of buffer QG were added to each gel band and the mixture was incubated on a thermomixer device at 50 °C and 700 rpm shaking for 20 min. Afterwards, 200 µl of Isopropanol were added to the mix containing precA (as precA is pretty short compared to the others, isopropanol is required). Afterwards, the whole reaction mix was loaded onto a gel extraction column and centrifuged for 1 min/max speed; flow-through was discarded. The column was subsequently washed with 500 µl of buffer QG and then 750 µl buffer PE, always centrifuging the column at max speed for 1 min at each washing step and discarding the flow-through. Finally, DNA was eluted in 20 µl of water.<br/>
| + | |
| - | <br/>
| + | |
| - | *<b>Concentration measurement of the pre-cut parts using Nanodrop</b>
| + | |
| - | | + | |
| - | <table style="margin:15px;border-collapse:collapse">
| + | |
| - | <tr><td style="padding:5px;border:2px solid black">Part</td><td style="padding:5px;border:2px solid black">DNA concentration</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">GFP</td><td style="padding:5px;border:1px solid black">12.5 ng/µl</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">LacZ</td><td style="padding:5px;border:1px solid black">15.11 ng/µl</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">RecA</td><td style="padding:5px;border:1px solid black">6.0 ng/µl</td></tr>
| + | |
| - | </table>
| + | |
| - | <html>
| + | |
| - | <h4><a style="color:#168dc4;" name="03/13/2012">03/13/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | *Ligation reactions were set up using 3-6 fold access of insert (precA) and 50 ng of each backbone (roughly).
| + | |
| - | ***precA-GFP: 4 µl (~50 ng) GFP-backbone + 1,5 µl (~9 ng) RecA + 2 µl T4 DNA ligase buffer + 11,5 µl water + 1 µl T4 ligase
| + | |
| - | ***precA-LacZ: 3 µl (~50 ng) LacZ-backbone + 1,5 µl (~9 ng) RecA + 2 µl T4 DNA ligase buffer + 12 µl water + 1 µl T4 ligase
| + | |
| - | ***psulA-GFP: 4 µl (~50 ng) GFP-backbone + 1 µl annealed SulA-oligos + 2 µl buffer + 12 µl water + 1 µl T4 ligase
| + | |
| - | ***psulA-LacZ: 3 µl (~50 ng) LacZ-backbone + 1 µl annealed SulA-oligos + 2 µl buffer + 13 µl water + 1 µl T4 ligase
| + | |
| - | **Incubation for 50 min at room temperature
| + | |
| - | **Heat inactivation of the T4 ligase at 70 °C for 5 min
| + | |
| - | *Afterwards, 10 µl of the ligation mix (chilled) were used for transforming 50 µl of <i>E. coli</i> Top10 chemocompetent cells.
| + | |
| - | <html>
| + | |
| - | <h4><a style="color:#168dc4;" name="03/14/2012">03/14/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | *<b>Screening of the four different biobrick clonings done by colony-PCR.</b><br/>
| + | |
| - | <b>Protocol:</b><br/>
| + | |
| - | A reaction mix was prepared containing 0.2 µl of each screening primer used for the screening, 9.6 µl of water and 10 µl of 2x PCR mastermix (fermentas). Finally, a colony was picked from a plate using a sterile pipette tip and was dipped into the PCR mix a few times. 7-8 clones were screened for each different cloning setup as follows:<br/>
| + | |
| - | **<u>psulA-GFP and psulA-LacZ:</u> primers psulA_fw and VR (standard sequencing primer) were used. Only in case psulA was successfully cloned into the backbone containing the reporter gene, we would get a PCR product. GFP would give a product of roughly 1000 bp, LacZ one of roughly 3500 bp.<br/>
| + | |
| - | **<u>precA-GFP and precA-LacZ:</u> primers VF2 and VR (standard sequencing primers) were used. We compared the product size of the different clones to the product size from the original vectors from the registry (only containing recA or LacZ). In case the cloning was successful, we should see a 200 bp shift in product size, which can be detected by gel electrophoresis.<br/>
| + | |
| - | The PCR program was done as follows:<br/>
| + | |
| - | <p style="font-family:monospace">94°C/3min||94°C/30s|60°C/30s|72°C/3min 45s||30x 72°C/10min|4°C/forever</p>
| + | |
| - | Afterwards, 3 ul PCR product were loaded onto a 1 % agarose gel.<br/>
| + | |
| - | <br/>
| + | |
| - | <html>
| + | |
| - | <br/>
| + | |
| - | <img class="gelimage" src="https://static.igem.org/mediawiki/2012hs/e/e6/RecA_GFP_gel.png" width ="500px"/><br/>
| + | |
| - | <b>Fig. 1 Colony-PCR screen of precA-GFP construct.</b> No detectable shift of any screened clone can be seen compared to the control. Screen has to be repeated.<br/>
| + | |
| - | <br/>
| + | |
| - | <img class="gelimage" src="https://static.igem.org/mediawiki/2012hs/2/23/RecA_LacZ_gel.png" width="500"/><br/>
| + | |
| - | <b>Fig. 2 Colony-PCR screen of precA-LacZ construct.</b> Most clones show a small shift upwards in product length. Furthermore, they show a second band at 1200 bp, probably due to unspecific primer binding.<br/>
| + | |
| - | <br/>
| + | |
| - | <img class="gelimage" src="https://static.igem.org/mediawiki/2012hs/b/b0/SulA_GFP-LacZ_gel.png"/><br/>
| + | |
| - | <b>Fig. 3 Colony-PCR screen of psulA-GFP (#3.1-3.8) and psulA-LacZ (#4.1-4.8).</b> Only clones containing the psulA promoter can give a PCR product of 1000 bp (psulA_GFP) or 3700 bp (psulA_LacZ). Positive clones are i.e. #3.4, #4.2, #4.8.<br/>
| + | |
| - | </html>
| + | |
| - | <br/>
| + | |
| - | The screening gave positive clones for all constructs (fig. 2, fig. 3), but not the precA_GFP construct (fig. 1). Therefore this screen has to be repeated using a larger number of colonies.<br/>
| + | |
| - | <br/>
| + | |
| - | *<b>5 ml LB-Amp overnight cultures were inoculated for clones #2.3, #3.4 and #4.8.</b>
| + | |
| - | <html>
| + | |
| - | <h4><a style="color:#168dc4;" name="03/15/2012">03/15/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | *The colony-PCR for the precA_GFP construct was repeated, now screening a number of 31 colonies. As control, again the amplicon of the original GFP biobrick was used.<br/>
| + | |
| - | <br/>
| + | |
| - | <html><img src="https://static.igem.org/mediawiki/2012hs/3/3f/RecA_GFP_rep_gel.png" /></html><br/>
| + | |
| - | <b>Fig. 1 Repetition of colony-PCR for precA_GFP construct.</b> Clones #6, 18 and 26 were positive (shift in amplicon length of ~200 bp).<br/>
| + | |
| - | <br/>
| + | |
| - | 3 positive colonies were detected, as they show a shift in band size of ~ 200 bp as expected by the introduction of the recA-promoter. Those were clones # 6, 18 and 26.<br/>
| + | |
| - | <br/>
| + | |
| - | *O/n LB-amp cultures were inoculated for clones #18 and #26.<br/>
| + | |
| - | <br/>
| + | |
| - | *Miniprep of the o/n cultures inoculated the day before for clones #2.3, 3.4 and 4.8 were done.<br/>
| + | |
| - | **concentrations measured using Nanodrop are all in between 70 and 140 ng/µl<br/>
| + | |
| - | <br/>
| + | |
| - | *Test digestions were performed in order to detect, whether the constructs would give the expected band patterns on the gel.
| + | |
| - | **<nowiki>#2.3 with EcoRI/BglI; as control the original LacZ biobrick was digested EcoRI/BgII as well</nowiki>
| + | |
| - | **<nowiki>#3.4 and #4.8 were digested NotI in order to see, whether an insert is present</nowiki><br/>
| + | |
| - | <br/>
| + | |
| - | <html><img src="https://static.igem.org/mediawiki/2012hs/0/01/Test_dig_1_gel.png" width=635 /></html><br/>
| + | |
| - | <b>Fig. 2 Test digestions for the constructs recA_LacZ (#2.3), psulA_GFP (#3.4) and sulA_LacZ (#4.8).</b> All constructs digested showed the right bands (left gel compared to virtual gel with expected bands on the right). Construct #2.3 gave the expected shift of the lowest band (arrow) compared to the LacZ-control construct (original biobrick BBa_K173004) due to the presence of the precA promoter.
| + | |
| - | <html>
| + | |
| - | <h4><a style="color:#168dc4;" name="03/16/2012">03/16/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | *Miniprep of precA_GFP clones #18 and #26 inoculated the previous
| + | |
| - | **Measurement of DNA concentration with the Nanodrop gave 71 ng/ul for both constructs<br/>
| + | |
| - | <br/>
| + | |
| - | *Test digestion of 1 ug of #18 and #26 with EcoRI/NcoI
| + | |
| - | **Band correct for both constructs
| + | |
| - | <html><img src="https://static.igem.org/mediawiki/2012hs/2/2b/Test_dig_2_gel.png" width="200"/></html><br/>
| + | |
| - | <br/>
| + | |
| - | *All constructs (clones #1.18, 2.3, 3.4, 4.8) were send for sequencing to GATC using standard biobrick sequencing primer VF2
| + | |
| - | <html>
| + | |
| - | <h4><a style="color:#168dc4;" name="03/17/2012">03/17/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | *Sequencing results were obtained from GATC for the first 4 Biobricks we did
| + | |
| - | **Sequence was confirmed for all 4 constructs; cloning was successful!
| + | |
| - | <table style="margin:15px;border-collapse:collapse">
| + | |
| - | <tr><td style="padding:5px;border:2px solid black">Part</td><td style="padding:5px;border:2px solid black">DNA Confirmation</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">RecA-GFP</td><td style="padding:5px;border:1px solid black">OK</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">RecA-LacZ</td><td style="padding:5px;border:1px solid black">OK</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">SulA-GFP</td><td style="padding:5px;border:1px solid black">OK</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">SulA-LacZ</td><td style="padding:5px;border:1px solid black">OK</td></tr>
| + | |
| - | </table>
| + | |
| - | <html>
| + | |
| - | <h4><a style="color:#168dc4;" name="05/08/2012">05/08/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | <b>Anealing of oligos for precB and precC:</b>
| + | |
| - | | + | |
| - | Got oligo sequences from <html><a href="http://www.ncbi.nlm.nih.gov/pubmed/">pubmed</a></html>
| + | |
| - | (looked for appropriate ATG and then took the following 70 basepairs as promoter sequence)
| + | |
| - | | + | |
| - | <b>RecB:</b><br/>
| + | |
| - | fwd.:
| + | |
| - | <p style="font-family:monospace;margin:10px">aattcgcggccgcttctagagCCTGAAGGCTGGAAAGTGTGGGAGAACGTCAGCGCGTTGCAGCAAACAATGCCCCTGATGAGTGAAAAGAc</p>
| + | |
| - | rev.:
| + | |
| - | <p style="font-family:monospace;margin:10px">ctaggTCTTTTCACTCATCAGGGGCATTGTTTGCTGCAACGCGCTGACGTTCTCCCACACTTTCCAGCCTTCAGGctctagaagcggccgcg</p>
| + | |
| - | | + | |
| - | | + | |
| - | <b>RecC:</b><br/>
| + | |
| - | fwd.:
| + | |
| - | <p style="font-family:monospace;margin:10px">aattcgcggccgcttctagagTTCACCCGGGGGCAGAGAAGGCGAGATGACCCGCCTGCATTGCCCGAATCGTCAGTAGTCAGGAGCCGCTc</p>
| + | |
| - | rev.:
| + | |
| - | <p style="font-family:monospace;margin:10px">ctaggAGCGGCTCCTGACTACTGACGATTCGGGCAATGCAGGCGGGTCATCTCGCCTTCTCTGCCCCCGGGTGAActctagaagcggccgcg</p>
| + | |
| - | | + | |
| - | | + | |
| - | Anealing-Protocol:
| + | |
| - | *5 ul of each oligo (diluted to a concentration of 100 mM)
| + | |
| - | *5 ul of NEB buffer 2
| + | |
| - | *35 ul of water
| + | |
| - | *Mix
| + | |
| - | *Heat up reaction mix to 95 °C for 5 minutes
| + | |
| - | *Cool down slowly to room temperature (then store at -20 °C)
| + | |
| - | | + | |
| - | | + | |
| - | <b>Ligation precB_LacZ / precC_lacZ:</b>
| + | |
| - | | + | |
| - | Add contents in the following order:
| + | |
| - | *12 ul water
| + | |
| - | *2 ul T4 DNA ligase buffer
| + | |
| - | *4 ul LacZ-backbone
| + | |
| - | *1 ul precB / precC
| + | |
| - | *1 ul T4 ligase
| + | |
| - | *leave 20 minutes at room temperature
| + | |
| - | *heat up to ca. 70°C to destroy ligase-enzymes
| + | |
| - | | + | |
| - | <b>Transformation of precB_LacZ / precC_LacZ into Top10:</b>
| + | |
| - | | + | |
| - | *Transformation-protocoll missing!
| + | |
| - | <html>
| + | |
| - | <h4><a style="color:#168dc4;" name="05/26/2012">05/26/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | <b>Colony PCR:</b><br/>
| + | |
| - | Screening of the two different biobrick clonings done by colony-PCR.
| + | |
| - | <br/><br/>
| + | |
| - | <b>Protocol:</b><br> A reaction mix was prepared containing 0.2 µl of each screening primer used for the screening, 9.6 µl of water and 10 µl of 2x PCR mastermix (fermentas). Finally, a colony was picked from a plate using a sterile pipette tip and was dipped into the PCR mix a few times. 12 clones were screened for each different cloning setup as follows:<br/>
| + | |
| - | <br/>
| + | |
| - | <u>precB_LacZ</u>: primers VF2: and precB Reverse: were used. Only in case precB was successfully cloned into the backbone containing the reporter gene, we would get a PCR product.
| + | |
| - | <br/><br/>
| + | |
| - | <u>precC_LacZ</u>: primers VF2 and VR (standard sequencing primers) were used. We compared the product size of the different clones to the product size from the original vectors from the registry (only containing precA or LacZ). In case the cloning was successful, we should see a 200 bp shift in product size, which can be detected by gel electrophoresis.
| + | |
| - | <br/><br/>
| + | |
| - | <b>The PCR program was done as follows:</b><br/>
| + | |
| - | 94°C/3min||94°C/30s|60°C/30s|72°C/3min 45s||30x 72°C/10min|4°C/forever
| + | |
| - | Afterwards, 3 µl PCR product were loaded onto a 1 % agarose gel.
| + | |
| - | | + | |
| - | <html>
| + | |
| - | <h4><a style="color:#168dc4;" name="05/28/2012">05/28/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | <b>Transferring the parts into standard registry plasmid pSB1C3:</b>
| + | |
| - | | + | |
| - | *Restriction digest of 1 µg of each sample and of the standard backbone
| + | |
| - | *After 30 min SapI is added to the backbone digest
| + | |
| - | *After overall 60 min Qiagen nucleotide removal kit was used to purify DNA
| + | |
| - | *heat inactivation of remaining enzymes at 80°C
| + | |
| - | *concentration measurement with nanodrop
| + | |
| - | *Ligation
| + | |
| - | <br/>
| + | |
| - | *heat inactivation at 70°C
| + | |
| - | | + | |
| - | <html>
| + | |
| - | <h4><a style="color:#168dc4;" name="06/02/2012-con">06/02/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | <b>Inoculate:</b><br/><br/>
| + | |
| - | <u>Protocol</u>: Constructs grown as overnightculture on agarplates:
| + | |
| - | <br/>
| + | |
| - | * #5 <html><a href="http://partsregistry.org/Part:BBa_K862000"> precA_LacZ</a></html>
| + | |
| - | * #7 <html><a href="http://partsregistry.org/Part:BBa_K862001"> psulA_LacZ</a></html>
| + | |
| - | * #8 <html><a href="http://partsregistry.org/Part:BBa_K862002"> precB_LacZ</a></html>
| + | |
| - | * #9 precC_LacZ<br/>
| + | |
| - | | + | |
| - | A reaction mix was prepared containing 20 ml LB medium and 20 µl Chloramphenicol (50mg/µl). Finally a single blue colony (except for #9) was picked from an agar plate using a sterile pipette tip and was dropped into a tube with 2 ml of the reaction mix.<br/>
| + | |
| - | Except for #9 we picked two colonies of each plate and inoculated separately. For plate #9, we picked four white colonies to check for basal expression.<br/>
| + | |
| - | After picking the colonies the tubes, in which the pipette tips where dropped, were put into 20 ml LB Medium .<br/>
| + | |
| - | Several hours later a miniprep of the grown cultures with the Qiagen Kit followed. DNA was eluted in TE Buffer.<br/>
| + | |
| - | Measurement of DNA concentration with Nanodrop:<br/>
| + | |
| - | <br/>
| + | |
| - | <table style="margin:15px;border-collapse:collapse">
| + | |
| - | <tr><td style="padding:5px;border:2px solid black">Part</td><td style="padding:5px;border:2px solid black">DNA concentration</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">#5.1</td><td style="padding:5px;border:1px solid black">16 ng/µl</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">#5.2</td><td style="padding:5px;border:1px solid black">6 ng/µl</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">#7.1</td><td style="padding:5px;border:1px solid black">46 ng/µl</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">#7.2</td><td style="padding:5px;border:1px solid black">57 ng/µl</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">#8.1</td><td style="padding:5px;border:1px solid black">84 ng/µl</td></tr>
| + | |
| - | <tr><td style="padding:5px;border:1px solid black">#8.2</td><td style="padding:5px;border:1px solid black">91 ng/µl</td></tr>
| + | |
| - | </table>
| + | |
| - | | + | |
| - | | + | |
| - | <html>
| + | |
| - | <h4><a style="color:#168dc4;" name="06/07/2012">06/07/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | <html>
| + | |
| - | <p><b>Three parts were sent to the parts registry</b>:
| + | |
| - | You can find all BioBricks under: <a href="http://2012HS.igem.org/Team:Heidelberg_LSL/Parts"> Parts </a> </p>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | <html>
| + | |
| - | <h2><a name="Calibration" style="color:#130ed2;">Calibration and Characterization</a></h2>
| + | |
| - | <br />
| + | |
| - | <h4><a name="04/27/2012">04/27/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | Transformation of <i>E.coli</i> strain BL21 (DE3) with our constructs the samples were plated on LB agarplates with Ampicillin.
| + | |
| - | | + | |
| - | <html>
| + | |
| - | <h4><a style="color:#130ed2;" name="04/28/2012">04/28/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | Inoculation of overnight cultures in LB liquid medium with Amp from the plates.
| + | |
| - | | + | |
| - | <html>
| + | |
| - | <h4><a style="color:#130ed2;" name="04/29/2012">04/29/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | <b> Experiment 1: </b><br/>
| + | |
| - | *day culture was inoculated with 1:30 dilution of overnight culture in LB with Ampicillin
| + | |
| - | *day culture was incubated for 3 h
| + | |
| - | *measurement of optical density using a photometer at 600 nm to confirm bacterial growth
| + | |
| - | *distribution of each 3 ml per sample to 6-well-plates
| + | |
| - | *7 replicate plates one per time span: 0 s, 5 s, 10 s, 30 s, 5 min, 10 min of exposure time
| + | |
| - | *Exposure to UV-light in the Intas gel IX imager with the above-mentioned time spans
| + | |
| - | *Addition of 30 µl X-Gal (2mg/ ml) to samples #5, #7 (constructs containing LacZ) plates sealed with parafilm
| + | |
| - | *Incubation at 37°C, 50 rpm for 45 min, visual color change: LacZ samples became blue
| + | |
| - | *measurement of optical density at 600 nm of #5 and #7<br/>
| + | |
| - | | + | |
| - | Problem: maximum absorption of X-Gal at 615-650 nm interferes with measurement of bacterial density at 600 nm – no change in absorption although blue colour was visible
| + | |
| - | <br/><br/>
| + | |
| - | <b> Experiment 2: </b><br/>
| + | |
| - | *overnight cultures centrifuged at 4000 rpm for 7 min
| + | |
| - | *pellet resuspended in 25 ml LB Amp
| + | |
| - | *The bacterial suspension is again exposed to UV-light with time spans of 0s, 20s, 60s, 300s, 600s.
| + | |
| - | *After the exposure X-Gal is added to the samples. Incubation at 37°C, 80 rpm
| + | |
| - | *first visible color change after 5 min in #7, #5 with X-Gal
| + | |
| - | *for a quantification the assays were plated out with duplicates on a 96w-Plate. LB medium was used as a blank reference. ONPG was added and the absorbance was measured with the plate reader
| + | |
| - | *ONPG was used because its absorbance maximum differs from the wavelength you use to measure the optical density
| + | |
| - | *after that we measured the optical density of our samples using the photometer to get the true expression.
| + | |
| - | | + | |
| - | <html>
| + | |
| - | <center><img src="https://static.igem.org/mediawiki/2012hs/8/80/Bildschirmfoto_2012-06-11_um_23.47.05.png" width="500"/> </center>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | | + | |
| - | <html>
| + | |
| - | <h4><a style="color:#130ed2;" name="05/27/2012">05/27/2012</a></h4>
| + | |
| - | </html>
| + | |
| - | | + | |
| - | Repetition of the second experiment at 29/04/2012.<br/>
| + | |
| | | | |
| - | *overnight cultures centrifuged at 4000 rpm for 7 min | + | <b>The following models have been created</b> |
| | + | * We should use these in the proposal, but continue to work on the Tinkercell model |
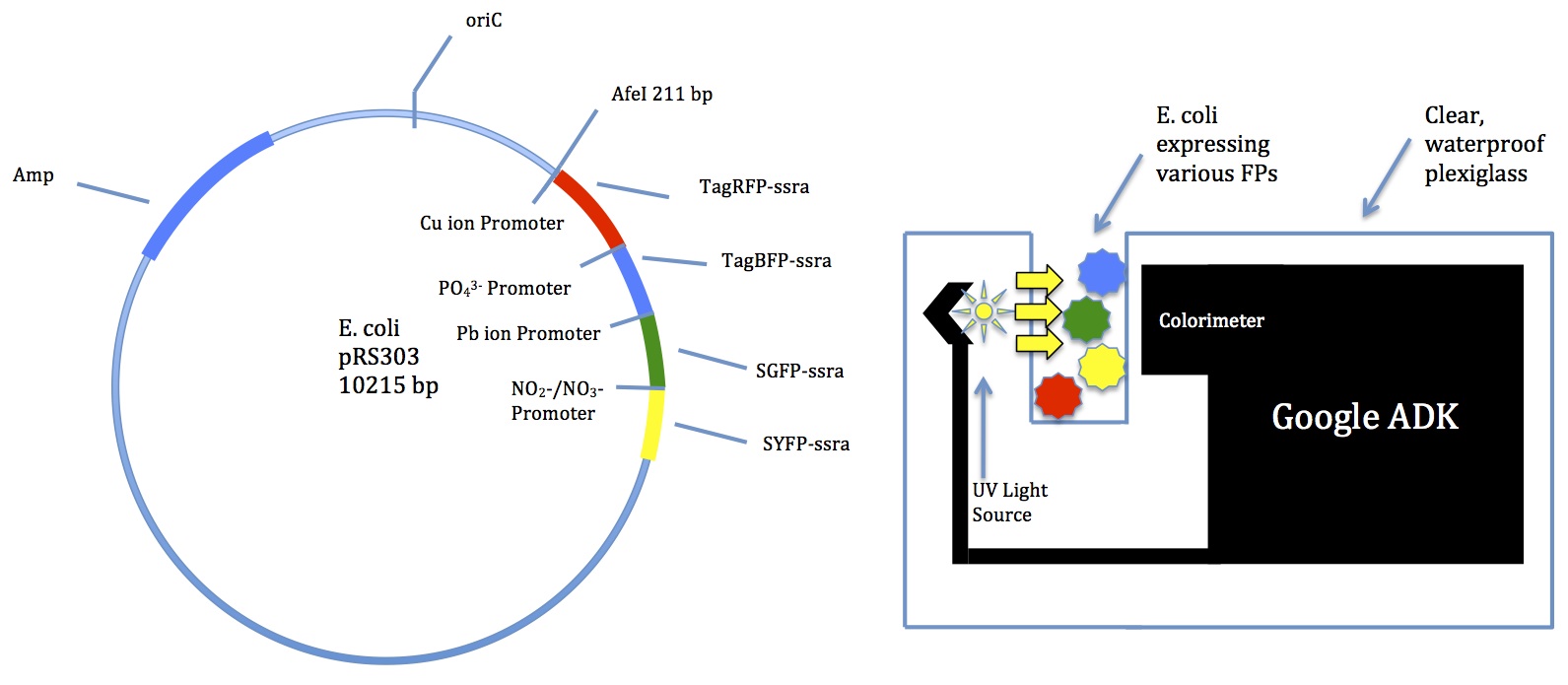
| | + | <div style="margin-top:7px;">[[Image:Design.jpg|center]]</div> |
| | | | |
| - | *pellet resuspended in 25 ml LB Amp<br/>
| + | <html><h4><a style="color:#00008B; float:right;" name="03/25/2013">03/25/2013</a></h4></html> |
| | | | |
| - | *The bacterial suspension is again exposed to UV-light with time spans of 10min 8min 6min 4min 2min 0min | + | <b>Editing poster on Google Doc</b> |
| | + | *Want to be finished by March 29th |
| | + | *Tinkercell model will probably not be ready in time |
| | + | **Having a lot of trouble getting fluorescent protein levels to zero when they should be |
| | + | **Danny will talk to Morgan to see if he can help |
| | + | *Edit Edit Edit! |
| | | | |
| - | *After the exposure X-Gal is added to the samples. Incubation at 37°C, 80rpm
| + | <html><h4><a style="color:#00008B; float:right;" name="03/27/2013">03/27/2013</a></h4></html> |
| | | | |
| - | <u>Experiment failed.</u><br/> | + | <b>Update on Proposal </b> |
| - | No clear graduation between the samples of different exposure times was observed.<br/>
| + | * Proposal is coming along well |
| - | <u>Possible causes</u>: big differences in incubation time after exposure<br/>
| + | * Need to finish the methods, but otherwise complete |
| | + | * Take pictures of the ADK and lab work |
| | | | |
| - | <b>Improvements in 4. experiment:</b><br/>
| + | <html><h4><a style="color:#00008B; float:right;" name="03/29/2013">03/29/2013</a></h4></html> |
| - | *24 wellplate recA and sulA
| + | |
| - | *for 5 min in UV light then the first row was transfered to another well-plate. The rest went again for 5 min under UV light, then the second row, with now an time period of 10 min.
| + | |
| - | *Same procedure for all the other rows. Time periods 5min 10min 15min 20min 30min and a controll sample with 0min
| + | |
| - | <html> | + | |
| - | <img src="https://static.igem.org/mediawiki/2012hs/7/7f/Bildschirmfoto_2012-06-12_um_00.03.28.png" width="635"/></html>
| + | |
| - | <br/>
| + | |
| - | <b>Fig. 1: Picture of the color gradient visible in the X-Gal assays with different exposure times</b>
| + | |
| - | <b>Testing of the precA_GFP construct</b><br/>
| + | |
| - | *Overnight culture of precA_GFP was diluted 1:5
| + | |
| - | *3ml of the culture were induced by UV radiation in the gel chamber for 30min
| + | |
| - | *The induced culture and 3ml of uninduced culture were incubated for 30min at 37°C / 80rpm
| + | |
| - | *After the incubation both cultures were compared using the fluorescence microscope
| + | |
| - | <br/>
| + | |
| - | <html>
| + | |
| - | <img src="https://static.igem.org/mediawiki/2012hs/6/6f/GFP_Measurement.PNG" width="635"/><br/>
| + | |
| - | <b>Fig. 2: Results of the GFP samples underneath fluorescence microscope</b>
| + | |
| - | <h4><a style="color:#130ed2;" name="05/28/2012-cal">05/28/2012</a></h4> | + | |
| - | </html> | + | |
| | | | |
| | + | <b>Proposal is completed </b> |
| | + | * Will submit soon |
| | | | |
| - | <b> Testing under real life conditions: </b> | + | <html><h4><a style="color:#00008B; float:right;" name="04/06/2013">04/06/2013</a></h4></html> |
| - | *culture of precA_LacZ and psulA_LacZ was centrifuged at 4000rpm for 8 min
| + | |
| - | *pellet was resuspended in LB Amp
| + | |
| - | *the samples were put into 2 6-well plates (3 ml/well)
| + | |
| - | *plates were tightly sealed, desinfected and put outdoor either into the sun or shadow for 75 min
| + | |
| - | *after 30 min incubation X-Gal was added to final concentration of 200 µg/ml the coloring of the samples were observed over time; intensity of the coloring in the wells were measured using the ImageJ software package
| + | |
| | | | |
| - | <html> | + | <b>Focus on creating the plasmid</b> |
| - | <img src="https://static.igem.org/mediawiki/2012hs/f/f9/Outdoor-Test.png" width=635> </img> | + | * Finished Tinkercell model will help a bunch |
| - | </html>
| + | * Do we want to think about ordering de novo from a gene synthesis company? |
| | + | * Computer modeling is the best approach for now |
| | | | |
| - | <html> | + | <html><h4><a style="color:#00008B; float:right;" name="04/17/2013">04/17/2013</a></h4></html> |
| - | <h4><a style="color:#130ed2;" name="06/07/2012-cal">06/07/2012</a></h4> | + | |
| - | </html> | + | |
| | | | |
| - | Overnight cultures precB_LacZ were diluted 1:2 with LB Medium and transferred onto 24-well-plates, 500 µl per well. Duplicates were exposed to UV-radiation in the UV-chamber for 5, 10, 15, 20 and 30 min. <br/>
| + | <b>Tinkercell Model </b> |
| - | X-Gal stock solution was diluted 1:100.<br/>
| + | * Danny has a working Tinkercell model for the copper detector! |
| - | After 1h incubation 50µl X-Gal was added to final concentration of 200 µg/ml. The coloring of the samples were observed over time. Intensity of the coloring in the wells were measured using the imageJ software package.<br/>
| + | **Outputs increased levels of TagRFP as ion concentration increases |
| | + | * We should be able to figure out the others soon |
| | + | * The lead detector may be especially difficult because the ion binds to a promoter to create a transcription factor |
| | | | |
| - | <html> | + | <html><h2><a name="Experimentation" style="color:#00008B;">Experimentation</a></h2> |
| - | <img src="https://static.igem.org/mediawiki/2012hs/c/c7/XGAL_Measurement.PNG" width="635" /><br/> | + | <h4><a style="color:#00008B; float:right;" name="04/27/2013">04/27/2013</a></h4></html> |
| - | <b>Fig. 1: All constructs in comparison</b> | + | |
| - | </div> | + | |
| - | </html> | + | |
| | | | |
| - | {{NotebookLower}}
| + | <b>Preliminary testing with the ADK </b> |
| - | /b> | + | * We want to work with the ADK for the next couple of weeks |
| - | 4 ml bacterial culture is pelleted by centrifugation at 10.000 rpm for 1 min. Afterwards, the pellet is resuspended in 250 µl buffer P1. 250 µl buffer P2 are added and the culture is gently inverted and incubated for 5 min. 350 µl buffer N3 are added and the suspension is centrifuged at 13.000 rpm for 10 min. The supernatant is subsequently loaded onto a miniprep column and centrifuged for 1 min/max speed. Afterwards the column is washed with buffer PB (500 µl) and buffer PE (750 µl) by loading onto the column and subsequent centrifugation for 1 min/max speed. After a last centrifugation step for drying the column (again 1 min/max speed), 50 µl of water are added in order to dissolve the DNA again. After 1 min incubation, DNA is eluted by centrifugation for 1 min/max speed.
| + | * Jack will attempt to code the ADK so it outputs to a text file |
| | + | * Potential preliminary tests |
| | + | ** Diluted glow sticks |
| | + | ** Dimmed LED lights |
| | + | ** pGLO E. coli |
| | | | |
| | + | <html><h4><a style="color:#00008B; float:right;" name="05/04/2013">05/04/2013</a></h4></html> |
| | | | |
| | + | <b>Upcoming ADK Testing</b> |
| | + | * The ADK has been coded |
| | + | * We have glow sticks, E. coli expressing GFP, and dim LEDs to test |
| | + | * Testing this Tuesday |
| | | | |
| | + | <html><h4><a style="color:#00008B; float:right;" name="05/07/2013">05/07/2013</a></h4></html> |
| | | | |
| - | ==Lab Notebook==
| + | <b>Data from the ADK preliminary experiment </b> |
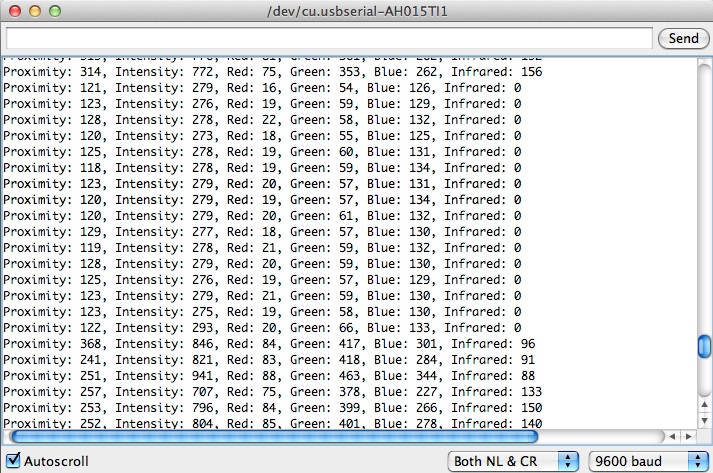
| - | To begin our experiment, we needed to create a device that could detect color. We chose to use the Google ADK, an open ended platform that allows users to take control of many sensors that come attached to an Arduino board, including a colorimeter. However, a standard ADK is set up to match any color presented to it, using LED lights. In some of our preliminary tests, the reflection of the light from these LEDs skewed data. Therefore, we modified the code of the ADK, using Google's software developer package, so that it does not turn on its LED lights but instead outputs colorimeter data to a computer. An example of this output is shown below.
| + | * Tested the sensing ability of the ADK today |
| | + | * Data is output in the form of the file below |
| | + | <div style="margin:10px 0px;">[[Image:Screenshot.jpg|700px|center]]</div> |
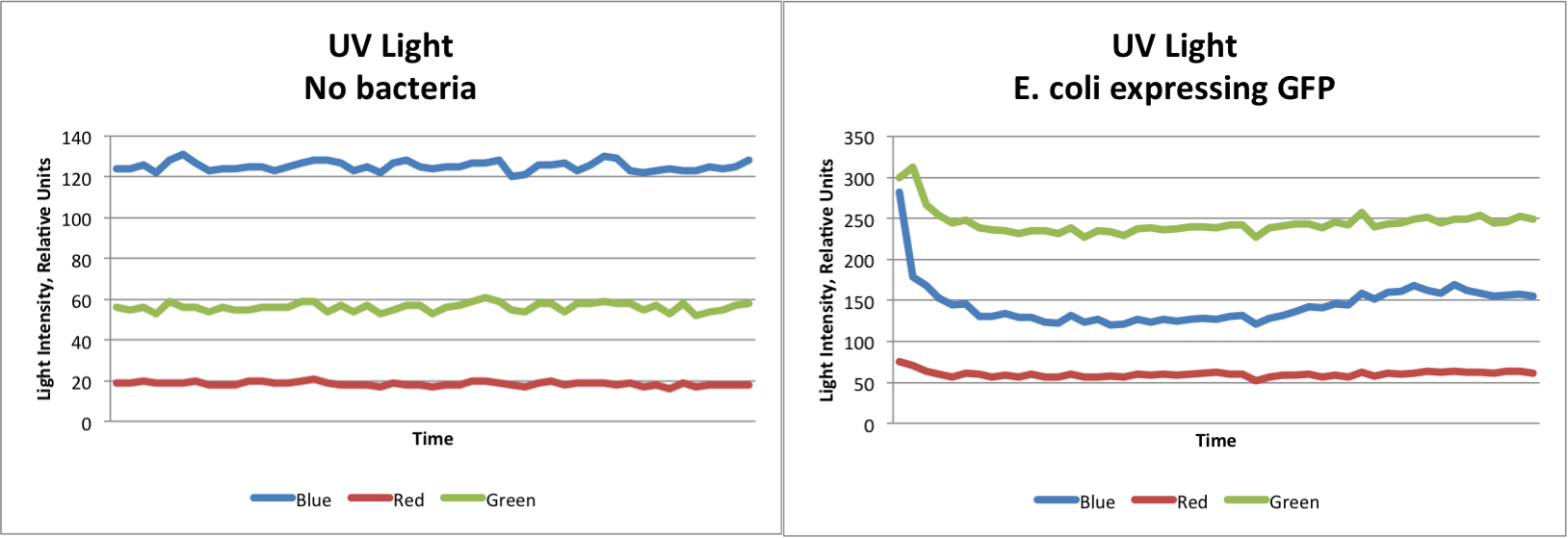
| | + | * The pGLO E. coli is the most closely related to our project and data is shown below |
| | + | <div style="margin:10px 0px;">[[Image:pGLOGraphs.png|center]]</div> |
| | + | * Data from LED lights is very similar but allowed us to understand the sensing "logic" of the ADK |
| | + | ** Showing red, green, or blue light increases only the color being shown |
| | + | ** Showing yellow light increases both red and green light |
| | + | ** Can identify which color is being shown from the data |
| | | | |
| - | [[File:Screenshot.jpg|400px]]
| + | <html><h4><a style="color:#00008B; float:right;" name="05/17/2013">05/17/2013</a></h4></html> |
| | | | |
| | + | <b>Completed Biobrick</b> |
| | + | * Everyone should begin working to finish their individual parts before the wiki freezes |
| | + | * Jack and Danny will take results from the Tinkercell model |
| | + | * Sequences for all parts have been found and the Biobrick as a whole is "completed" |
| | + | * Meet next week to start uploading everything to the wiki and start to design the poster |
| | | | |
| - | As a foray into our project, we first took some preliminary data using our modified Google ADK and pGLO E. coli expressing Green Fluorescent Protein. We found that we were able to detect a distinct change in color. Some baseline color was present from the UV Light source used for the experiment. The graphs below clearly show that our system is able to detect E. coli expressing GFP.
| + | <html><h4><a style="color:#00008B; float:right;" name="06/06/2013">06/06/2013</a></h4></html> |
| | | | |
| - | [[File:pGLOGraphs.png|800px]] | + | <b>Experimentation Data</b> |
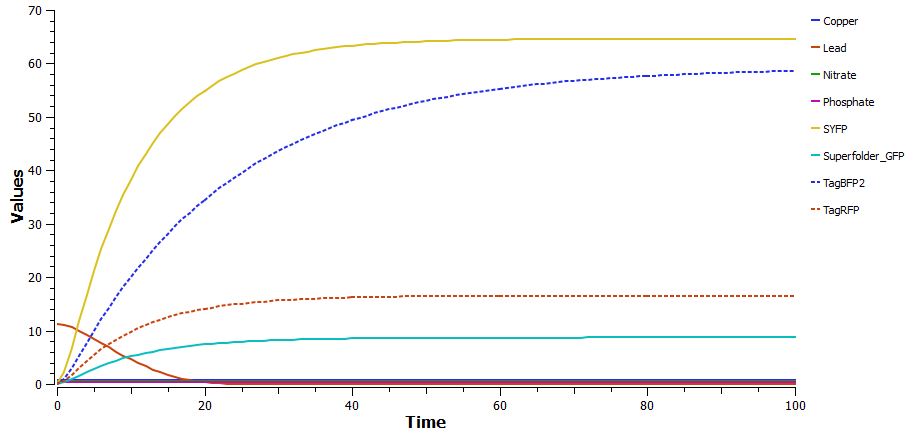
| | + | * Poster abstract, introduction, and methods are completed |
| | + | * The Tinkercell data shows our part effectively responds to the influx of the ions for which we are testing |
| | + | [[Image:Results.jpg|575|center]] |
| | + | * We will write up the results in the coming week |
| | | | |
| - | Small LED lights were used to test the ADK's ability to detect red, blue, and yellow lights. The results showed the same trends as the GFP test, indicating that we would be able to distinguish between several different colors expressed. In general, red, green, and blue lights show dramatic increases in the intensity of that color, while the other two colors remain about the same. Yellow light however, increase both red and green light, while blue remains at the same level. From these observations, we are able to identify which type of light is shown based only on the data collected.
| + | <html><h4><a style="color:#00008B; float:right;" name="06/15/2013">06/15/2013</a></h4></html> |
| - | {{clear}}
| + | |
| | | | |
| | + | <b>Poster Update</b> |
| | + | * Draft poster is complete! |
| | + | [[Image:Poster.jpg|475|center]] |
| | + | * Will continue to edit |
| | + | * Need to start adding all of our stuff to the wiki |
| | }} | | }} |
 "
"