Team:BV CAPS Kansas/Project/Methods
From 2013hs.igem.org
(Difference between revisions)
Meaton2222 (Talk | contribs) |
Meaton2222 (Talk | contribs) |
||
| (25 intermediate revisions not shown) | |||
| Line 714: | Line 714: | ||
position: relative; | position: relative; | ||
width: 208px; | width: 208px; | ||
| - | margin-top: | + | margin-top:8px; |
float: right; | float: right; | ||
height: auto; | height: auto; | ||
| Line 825: | Line 825: | ||
border: 1px solid #FFFFFF; | border: 1px solid #FFFFFF; | ||
} | } | ||
| - | + | <center> | |
#tweets { | #tweets { | ||
| Line 911: | Line 911: | ||
</head> | </head> | ||
| - | + | </center> | |
<body class="mediawiki ltr ns-0 ns-subject page-Team_UC_Davis"> | <body class="mediawiki ltr ns-0 ns-subject page-Team_UC_Davis"> | ||
| Line 949: | Line 949: | ||
<li ><a href="https://2013hs.igem.org/Team:BV_CAPS_Kansas/Gallery" | <li ><a href="https://2013hs.igem.org/Team:BV_CAPS_Kansas/Gallery" | ||
| - | title="Fun"><b>Fun</b></a></li> | + | title="Fun"><b>Fun</b></a> <ul> |
| + | <li ><a title="Facebook">Facebook</a> | ||
| + | <ul> | ||
| + | <li ><a href="https://www.facebook.com/groups/368737559905193/?fref=ts">HS iGEM</a></li> | ||
| + | <li ><a href="https://www.facebook.com/capsigem2013?fref=ts">CAPS iGEM</a></li> | ||
| + | <li ><a href="https://www.facebook.com/groups/igemmers/?fref=ts">iGEM</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li ><a title="Twitter">Twitter</a> | ||
| + | <ul> | ||
| + | <li ><a href="https://twitter.com/CAPSiGEM">CAPS iGEM</a></li> | ||
| + | <li ><a href="https://twitter.com/iGEM">iGEM HQ</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li ><a title="Slideshows">Slideshows</a> | ||
| + | <ul> | ||
| + | <li ><a href="https://static.igem.org/mediawiki/2013hs/b/bd/CAPS_iGEM_Slideshow_2013.pdf">2013</a></li> | ||
| + | <li ><a href="https://static.igem.org/mediawiki/2013hs/6/66/IGEM_2012_Slideshow.pdf">2012</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li ><a href="https://2013hs.igem.org/Team:BV_CAPS_Kansas/Gallery">Glycolysis Poem</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | |||
| + | <ul> | ||
| + | <li ><a href="https://2013hs.igem.org/Team:BV_CAPS_Kansas/Gallery">Glycolysis Poem</a></li> | ||
| + | </ul> | ||
| + | </li></li> | ||
<li ><a href="https://2013hs.igem.org/Team:BV_CAPS_Kansas/Safety" | <li ><a href="https://2013hs.igem.org/Team:BV_CAPS_Kansas/Safety" | ||
| Line 1,038: | Line 1,066: | ||
</div> | </div> | ||
<div id="myleftrightbox" class="fourboxes2-4"> | <div id="myleftrightbox" class="fourboxes2-4"> | ||
| - | <a href="https://2013hs.igem.org"><img src="https://static.igem.org/mediawiki/2013hs/9/9f/IGEM_Brick_Image.jpg"></a> | + | <a href="https://2013hs.igem.org/Jamboree"><img src="https://static.igem.org/mediawiki/2013hs/9/9f/IGEM_Brick_Image.jpg"></a> |
</div> | </div> | ||
</div> | </div> | ||
| Line 1,102: | Line 1,130: | ||
</center> | </center> | ||
| + | <center> | ||
| + | <a href="https://2008.igem.org/Team:Hawaii" target="_blank"><img src="https://static.igem.org/mediawiki/2013hs/6/6a/Hawaiilogo.jpg" width="200"></a> | ||
| + | </center> | ||
<center> | <center> | ||
<a href="https://2012.igem.org/Team:UC_Davis" target="_blank"><img src="https://static.igem.org/mediawiki/2011/4/40/UCD_BME_logo_minimal_copy.png" width="200 height="70"></a> | <a href="https://2012.igem.org/Team:UC_Davis" target="_blank"><img src="https://static.igem.org/mediawiki/2011/4/40/UCD_BME_logo_minimal_copy.png" width="200 height="70"></a> | ||
| Line 1,131: | Line 1,162: | ||
<div id="myleftbox" class="smallbox"> | <div id="myleftbox" class="smallbox"> | ||
<h1>Methods</h1> | <h1>Methods</h1> | ||
| - | + | Inserting the BBa_K977000 pyruvate kinase gene into BBa_K125000 vector required a number of different steps and techniques.<br><br> | |
| - | + | ||
| - | < | + | Because the vector already contained a usable promoter, the main procedure involved four different methods.<br> |
| - | + | ||
| - | < | + | <ol> |
| - | + | 1. Using site-directed mutagenesis to remove a naturally-occurring PstI restriction site in the pyruvate kinase gene.<br> | |
| - | <br> | + | 2. Using PCR to “flank” the pyruvate kinase gene with the BioBrick Standard Restiction Enzyme sites.<br> |
| - | + | 3. Using 3A Assembly to insert the new part into the BBa_K125000 vector.<br> | |
| - | + | 4. Transforming the cyanobacteria.</ol><br> | |
| - | < | + | |
| - | + | <b>Site-Directed Mutagenesis</b><br><br> | |
| - | + | Site-directed mutagenesis is a method for altering the sequence of DNA and can be used to produce both point and frame-shift mutations. In this case, it was possible to take advantage of codon redundancy and alter the DNA sequence without altering the amino acid sequence. It was necessary to do sitedirect mutagenesis because the pyruvate kinase gene contains in its sequence two PstI restriction sites. (CTGCAG)<br><br> | |
| - | + | ||
| - | Site-directed mutagenesis is a method for altering the sequence of DNA and can be used to | + | Because PSTI is part of biobrick assembly, it was necessary to use site-directed mutagenesis to remove these sites prior to cleaving and ligating the part. Otherwise PstI would chop the gene to pieces whenever we (or anybody else) tried to assemble anything with it.<br><br> |
| - | Primer design was | + | |
| + | Primer design was very simple, as the primers were identical to the gene except that they contained the desired mutations. Again, the primers were about 20 bp to ensure that they would only attach to one region along the gene. | ||
| + | <br><br> | ||
| + | |||
| + | <center><a ><img src="https://static.igem.org/mediawiki/2013hs/e/e1/Site_Directed_Mutagenesis.jpg" width="400" title="Expand"></a></center><br> | ||
<i>Forward primer</i><br> | <i>Forward primer</i><br> | ||
Template strand- 5’- ctcatctcc<font color="red">ctgcag</font>gtgaagcag 3’<br> | Template strand- 5’- ctcatctcc<font color="red">ctgcag</font>gtgaagcag 3’<br> | ||
| Line 1,151: | Line 1,186: | ||
Primer -5’- ctcatctcc<font color="red">ct</font><font color="blue">c</font><font color="red">cag</font>gtgaagcag 3’<br> | Primer -5’- ctcatctcc<font color="red">ct</font><font color="blue">c</font><font color="red">cag</font>gtgaagcag 3’<br> | ||
(Restriction sites are in red, point mutation is in blue) | (Restriction sites are in red, point mutation is in blue) | ||
| - | <br> | + | <br><br> |
<i>Reverse primer</i><br> | <i>Reverse primer</i><br> | ||
Again, the reverse primer necessitated the use of the reverse compliment of the original sequence. <br> | Again, the reverse primer necessitated the use of the reverse compliment of the original sequence. <br> | ||
Template strand 5’- CTGCTTCACCTGCAGGGAGATGAG -3’<br> | Template strand 5’- CTGCTTCACCTGCAGGGAGATGAG -3’<br> | ||
Primer 5’- CTGCTTCACCTGCAGGGAGATGAG 3’ | Primer 5’- CTGCTTCACCTGCAGGGAGATGAG 3’ | ||
| - | <br> | + | <br><br> |
| - | + | ||
| - | <b> | + | <a href="http://parts.igem.org/Help:Constructing_Parts?title=Help:Constructing_Parts">Click here</a> for the official Standard Registry Procedures regarding site-directed mutagenisis.<br><br> |
| - | <br> | + | |
| - | <br> | + | <b>PCR</b><br> |
| + | In order to get the pyruvate kinase gene to conform to Biobrick Standards, it was necessary to add the four Biobrick restriction sites.<br><br> | ||
| + | |||
| + | This was done with a variation on PCR annealing and extension per the instructions given by the Registry. Theoretically, this method was simple. All it required was forwards and reverse primers that consisted of the BioBrick prefixes and suffixes and the first twenty base pairs of the gene.<br><br> | ||
| + | |||
| + | <i>Forward primer</i><br> | ||
| + | 5’-<font color="red"> GTTTCTTGGAATTCGCGGCCGCTTCTAGATG </font> <font color="green"> TCAAAGTCCCACAGTGAAGC</font> -3’ | ||
| + | <br><br><i>Reverse Primer</i><br> | ||
| + | To orient the sequence 5’-to-3’, it was necessary to use the reverse compliment of the last twenty base pairs of the gene. <br> | ||
| + | 5’-<font color="red"> GTTTCTTCCTGCAGCGGCCGCTACTAGTA </font> <font color="green"> TTATTACGGGACGGGCACCACGCGCA </font> -3’ | ||
| + | <br>(Red is the tails, green is the gene) <br><br> | ||
| + | After this, it would be possible to proceed with PCR as normal, and though the region of restriction sites would “hang off” in the first round of PCR, they would be added to the gene as “flanks” in subsequent rounds as the extension stage would transcribe the flanks on as well as the gene’s DNA. However, a number of challenges presented themselves.<br><br> | ||
| + | |||
| + | <center><a ><img src="https://static.igem.org/mediawiki/2013hs/a/a7/Primer_for_Flanks.png" width="400" title="Expand"></a></center><br> | ||
| + | |||
| + | At 51 and 55 bp, the length of the primers far exceeded the typical 40-45 bp. The long “free ends” that contained the restriction sites threatened to rip the section of matching sequence away from the template strand. It proved impossible to shorten these primers while still including the standard restriction site. Nonetheless, the primers were ordered and the experiment was attempted. After running the results on a gel, the length of the resulting fragments suggested that the PCR was successful.<br><br> | ||
| + | |||
| + | |||
| + | |||
| + | As for the exact procedure, the <a href="http://parts.igem.org/Help:Protocol/PCR_Standardization">Procedure</a> offered by the registry was used. <br><br> | ||
| + | |||
| + | Upon creating the part, we used 3A assembly to insert the part into the BBa_K997000 vector (It’s completely standard, so <a href="http://parts.igem.org/Help:Protocol/3A_Assembly">look it up!</a>) and <a href="http://parts.igem.org/Help:Protocols/Transformation">transformed</a> the cyanobacteria.<br><br> | ||
| + | |||
| + | In the future, we might repeat this experiment with yeast, and though the sequences would be different the procedure would be essentially the same.<br><br> | ||
| + | |||
| + | <b>Other Standard iGEM Protocols for Preparing the our Vector and Tranforming our Part into our Cyanobacteria Chassis</b><ol> | ||
| + | <li><a href="http://parts.igem.org/Help:Protocols/Competent_Cells">Preparation of Competent Cells</a></li> | ||
| + | <li><a href="http://parts.igem.org/Help:Protocols/Transformation">Transformation</a></li> | ||
| + | <li><a href="http://parts.igem.org/Help:Protocols/Miniprep">Plasmid Mini Prep</a></li> | ||
| + | <li><a href="http://parts.igem.org/Help:Protocols/Restriction_Digest">Restiction Digest</a></li> | ||
| + | <li><a href="http://parts.igem.org/Help:Protocols/Ligation">Ligation</a></li> | ||
| + | <li><a href="http://parts.igem.org/Help:Assembly/3A_Assembly">3A Assembly</a></li></ol> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | <b>Tinker Cell</b><br> | ||
| + | |||
| + | A program called TinkerCell was utilized to help visualize the steps taken through glycolysis and the fatty-acid pathway to create alkanes. The image created in TinkerCell allows one to see each step in the modification of a molecule. Beginning with glucose and ending with alkanes, the image shows what the molecule becomes, which enzyme catalyzes the reaction, what type of reaction occurs, what is required for the reaction to occur, and the other products of the reaction.<br><br> | ||
| + | |||
| + | <b>Asssay for Pyruvate Kinase Activity</b><br> | ||
| + | |||
| + | Photosynthetic microbes, like cyanobacteria, hold the potential to produce biofuel with only sunlight and adequate growing conditions. These microbes do not need much space or require nutrient input to create fuel. However, biofuel output through this method is relatively low, which sparked our interest.<br><br> | ||
| + | <center><a href="https://static.igem.org/mediawiki/2013hs/7/7f/Other_Metabolilsm_Image.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2013hs/7/7f/Other_Metabolilsm_Image.jpg" title="Expand" width="400"></a></center><br> | ||
| + | Whether it is diesel fuel or gasoline, most of the energy comes from alkanes. A cell makes alkanes by the fatty-acid pathway. A main resource needed for the fatty-acid pathway, acetyl-CoA, is produced by glycolysis. We hypothesize that if we increase the resources required for the production of alkanes, then the cell will produce more alkanes.<br><br> | ||
| + | By overexpressing the catalytic protein pyruvate kinase, we hope to increase pyruvate within the cell. This should allow for an increase in activity within the alkane fatty-acid pathway because of pyruvates conversion to acetyl-CoA.<br><br> | ||
| + | <ol>PEP phosphorylates ADP 1 molecule pyruvate + 1 molecule ATP</ol><br> | ||
| + | To determine the amount of pyruvate activity produced by our cyanobacteria (engineered for overexpression of pyruvate kinase), we must assay for pyruvate kinase. A cocktail of magnesium chloride, ADP, EDTA, bicine (buffer), NADH and lactate dehydrogenase is made. Added to the purified pyruvate from the cyanobacteria plus PEP, the lactate dehydrogenase will utilize the NADH to reduce pyruvate to lactate. The amount of lactate produced indicates the output of pyruvate from the cell and can be read using a spectrophotometer at 340 nm. Using the spectrophotometer readings and a mathematical formula that takes into account dissociation rate of PEP into pyruvate into lactate acid, our initial pyruvate amount is determined. | ||
| + | <br><br> | ||
| + | |||
| + | <b>Assay for Alkanes</b><br> | ||
| + | |||
| + | Whether it is diesel fuel or gasoline, most of the energy comes from alkanes. A cell makes alkanes by the fatty-acid pathway. A main resource needed for the fatty-acid pathway, acetyl-CoA, is produced by glycolysis from pyruvate. <a href="https://2013hs.igem.org/Team:BV_CAPS_Kansas/Solutions#pathway">Click here</a> to review the alkane pathway. We hypothesize that if we increase the resources required for the production of alkanes, then the cell will produce more alkanes.<br><br> | ||
| + | Once it is determined that we have sufficient pyruvate production from our cyanobacteria (as determined by the pyruvate kinase assay), we will do lipid profiling by gas chromatography to verify the lipid produced is the substrate needed for the cell to produce alkanes. Gas chromatography takes the initial compound we wish to analyze (lipid produced from our cyanobacteria with over-expression of pyruvate kinase) and binds it to the liquid on the walls of the gas chromatography column. The lipid reacts with an added inert gas. This process yields a peak in the spectrum specific to the lipid in question. We anticipate that the lipid produced by our cyanobacteria will be the substrate for the cell to undergo the fatty-acid pathway to produce alkanes.<br><br> | ||
| - | + | The next step will be to confirm that alkane production has been completed by the cell. Working with our mentor, Dr. Fenton, we will also use gas chromatography to verify the alkane produced. | |
| - | |||
</div> | </div> | ||
Latest revision as of 02:03, 22 June 2013

BV CAPS iGEM Tweets
Methods
Inserting the BBa_K977000 pyruvate kinase gene into BBa_K125000 vector required a number of different steps and techniques.Because the vector already contained a usable promoter, the main procedure involved four different methods.
-
1. Using site-directed mutagenesis to remove a naturally-occurring PstI restriction site in the pyruvate kinase gene.
2. Using PCR to “flank” the pyruvate kinase gene with the BioBrick Standard Restiction Enzyme sites.
3. Using 3A Assembly to insert the new part into the BBa_K125000 vector.
4. Transforming the cyanobacteria.
Site-Directed Mutagenesis
Site-directed mutagenesis is a method for altering the sequence of DNA and can be used to produce both point and frame-shift mutations. In this case, it was possible to take advantage of codon redundancy and alter the DNA sequence without altering the amino acid sequence. It was necessary to do sitedirect mutagenesis because the pyruvate kinase gene contains in its sequence two PstI restriction sites. (CTGCAG)
Because PSTI is part of biobrick assembly, it was necessary to use site-directed mutagenesis to remove these sites prior to cleaving and ligating the part. Otherwise PstI would chop the gene to pieces whenever we (or anybody else) tried to assemble anything with it.
Primer design was very simple, as the primers were identical to the gene except that they contained the desired mutations. Again, the primers were about 20 bp to ensure that they would only attach to one region along the gene.

Forward primer
Template strand- 5’- ctcatctccctgcaggtgaagcag 3’
Primer -5’- ctcatctccctccaggtgaagcag 3’
(Restriction sites are in red, point mutation is in blue)
Reverse primer
Again, the reverse primer necessitated the use of the reverse compliment of the original sequence.
Template strand 5’- CTGCTTCACCTGCAGGGAGATGAG -3’
Primer 5’- CTGCTTCACCTGCAGGGAGATGAG 3’
Click here for the official Standard Registry Procedures regarding site-directed mutagenisis.
PCR
In order to get the pyruvate kinase gene to conform to Biobrick Standards, it was necessary to add the four Biobrick restriction sites.
This was done with a variation on PCR annealing and extension per the instructions given by the Registry. Theoretically, this method was simple. All it required was forwards and reverse primers that consisted of the BioBrick prefixes and suffixes and the first twenty base pairs of the gene.
Forward primer
5’- GTTTCTTGGAATTCGCGGCCGCTTCTAGATG TCAAAGTCCCACAGTGAAGC -3’
Reverse Primer
To orient the sequence 5’-to-3’, it was necessary to use the reverse compliment of the last twenty base pairs of the gene.
5’- GTTTCTTCCTGCAGCGGCCGCTACTAGTA TTATTACGGGACGGGCACCACGCGCA -3’
(Red is the tails, green is the gene)
After this, it would be possible to proceed with PCR as normal, and though the region of restriction sites would “hang off” in the first round of PCR, they would be added to the gene as “flanks” in subsequent rounds as the extension stage would transcribe the flanks on as well as the gene’s DNA. However, a number of challenges presented themselves.

At 51 and 55 bp, the length of the primers far exceeded the typical 40-45 bp. The long “free ends” that contained the restriction sites threatened to rip the section of matching sequence away from the template strand. It proved impossible to shorten these primers while still including the standard restriction site. Nonetheless, the primers were ordered and the experiment was attempted. After running the results on a gel, the length of the resulting fragments suggested that the PCR was successful.
As for the exact procedure, the Procedure offered by the registry was used.
Upon creating the part, we used 3A assembly to insert the part into the BBa_K997000 vector (It’s completely standard, so look it up!) and transformed the cyanobacteria.
In the future, we might repeat this experiment with yeast, and though the sequences would be different the procedure would be essentially the same.
Other Standard iGEM Protocols for Preparing the our Vector and Tranforming our Part into our Cyanobacteria Chassis
- Preparation of Competent Cells
- Transformation
- Plasmid Mini Prep
- Restiction Digest
- Ligation
- 3A Assembly
Tinker Cell
A program called TinkerCell was utilized to help visualize the steps taken through glycolysis and the fatty-acid pathway to create alkanes. The image created in TinkerCell allows one to see each step in the modification of a molecule. Beginning with glucose and ending with alkanes, the image shows what the molecule becomes, which enzyme catalyzes the reaction, what type of reaction occurs, what is required for the reaction to occur, and the other products of the reaction.
Asssay for Pyruvate Kinase Activity
Photosynthetic microbes, like cyanobacteria, hold the potential to produce biofuel with only sunlight and adequate growing conditions. These microbes do not need much space or require nutrient input to create fuel. However, biofuel output through this method is relatively low, which sparked our interest.
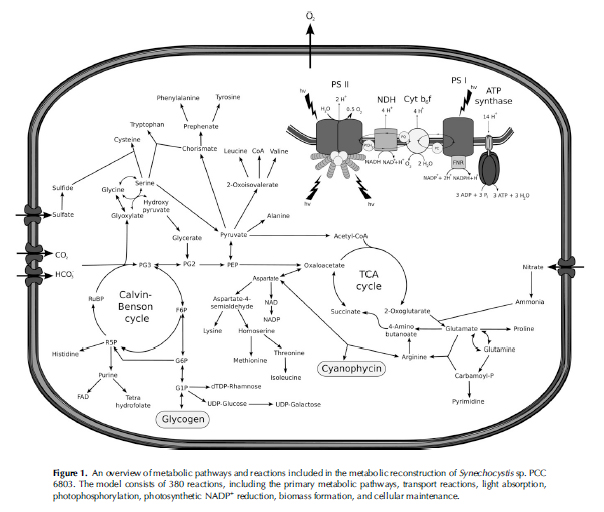
Whether it is diesel fuel or gasoline, most of the energy comes from alkanes. A cell makes alkanes by the fatty-acid pathway. A main resource needed for the fatty-acid pathway, acetyl-CoA, is produced by glycolysis. We hypothesize that if we increase the resources required for the production of alkanes, then the cell will produce more alkanes.
By overexpressing the catalytic protein pyruvate kinase, we hope to increase pyruvate within the cell. This should allow for an increase in activity within the alkane fatty-acid pathway because of pyruvates conversion to acetyl-CoA.
- PEP phosphorylates ADP 1 molecule pyruvate + 1 molecule ATP
To determine the amount of pyruvate activity produced by our cyanobacteria (engineered for overexpression of pyruvate kinase), we must assay for pyruvate kinase. A cocktail of magnesium chloride, ADP, EDTA, bicine (buffer), NADH and lactate dehydrogenase is made. Added to the purified pyruvate from the cyanobacteria plus PEP, the lactate dehydrogenase will utilize the NADH to reduce pyruvate to lactate. The amount of lactate produced indicates the output of pyruvate from the cell and can be read using a spectrophotometer at 340 nm. Using the spectrophotometer readings and a mathematical formula that takes into account dissociation rate of PEP into pyruvate into lactate acid, our initial pyruvate amount is determined.
Assay for Alkanes
Whether it is diesel fuel or gasoline, most of the energy comes from alkanes. A cell makes alkanes by the fatty-acid pathway. A main resource needed for the fatty-acid pathway, acetyl-CoA, is produced by glycolysis from pyruvate. Click here to review the alkane pathway. We hypothesize that if we increase the resources required for the production of alkanes, then the cell will produce more alkanes.
Once it is determined that we have sufficient pyruvate production from our cyanobacteria (as determined by the pyruvate kinase assay), we will do lipid profiling by gas chromatography to verify the lipid produced is the substrate needed for the cell to produce alkanes. Gas chromatography takes the initial compound we wish to analyze (lipid produced from our cyanobacteria with over-expression of pyruvate kinase) and binds it to the liquid on the walls of the gas chromatography column. The lipid reacts with an added inert gas. This process yields a peak in the spectrum specific to the lipid in question. We anticipate that the lipid produced by our cyanobacteria will be the substrate for the cell to undergo the fatty-acid pathway to produce alkanes.
The next step will be to confirm that alkane production has been completed by the cell. Working with our mentor, Dr. Fenton, we will also use gas chromatography to verify the alkane produced.
 "
"














